Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains
- PMID: 18559114
- PMCID: PMC2443715
- DOI: 10.1186/1471-2148-8-176
Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains
Abstract
Background: C2H2 zinc finger genes (C2H2-ZNF) constitute the largest class of transcription factors in humans and one of the largest gene families in mammals. Often arranged in clusters in the genome, these genes are thought to have undergone a massive expansion in vertebrates, primarily by tandem duplication. However, this view is based on limited datasets restricted to a single chromosome or a specific subset of genes belonging to the large KRAB domain-containing C2H2-ZNF subfamily.
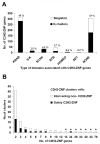
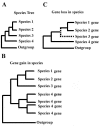
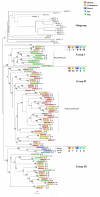
Results: Here, we present the first comprehensive study of the evolution of the C2H2-ZNF family in mammals. We assembled the complete repertoire of human C2H2-ZNF genes (718 in total), about 70% of which are organized into 81 clusters across all chromosomes. Based on an analysis of their N-terminal effector domains, we identified two new C2H2-ZNF subfamilies encoding genes with a SET or a HOMEO domain. We searched for the syntenic counterparts of the human clusters in other mammals for which complete gene data are available: chimpanzee, mouse, rat and dog. Cross-species comparisons show a large variation in the numbers of C2H2-ZNF genes within homologous mammalian clusters, suggesting differential patterns of evolution. Phylogenetic analysis of selected clusters reveals that the disparity in C2H2-ZNF gene repertoires across mammals not only originates from differential gene duplication but also from gene loss. Further, we discovered variations among orthologs in the number of zinc finger motifs and association of the effector domains, the latter often undergoing sequence degeneration. Combined with phylogenetic studies, physical maps and an analysis of the exon-intron organization of genes from the SCAN and KRAB domains-containing subfamilies, this result suggests that the SCAN subfamily emerged first, followed by the SCAN-KRAB and finally by the KRAB subfamily.
Conclusion: Our results are in agreement with the "birth and death hypothesis" for the evolution of C2H2-ZNF genes, but also show that this hypothesis alone cannot explain the considerable evolutionary variation within the subfamilies of these genes in mammals. We, therefore, propose a new model involving the interdependent evolution of C2H2-ZNF gene subfamilies.
Figures


Similar articles
-
The ancient mammalian KRAB zinc finger gene cluster on human chromosome 8q24.3 illustrates principles of C2H2 zinc finger evolution associated with unique expression profiles in human tissues.BMC Genomics. 2010 Mar 26;11:206. doi: 10.1186/1471-2164-11-206. BMC Genomics. 2010. PMID: 20346131 Free PMC article.
-
A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors.Genome Res. 2006 May;16(5):669-77. doi: 10.1101/gr.4842106. Epub 2006 Apr 10. Genome Res. 2006. PMID: 16606702 Free PMC article.
-
Deep vertebrate roots for mammalian zinc finger transcription factor subfamilies.Genome Biol Evol. 2014 Mar;6(3):510-25. doi: 10.1093/gbe/evu030. Genome Biol Evol. 2014. PMID: 24534434 Free PMC article.
-
Evolution of KRAB-containing zinc finger proteins and their roles in species evolution.Yi Chuan. 2016 Nov 20;38(11):971-978. doi: 10.16288/j.yczz.16-056. Yi Chuan. 2016. PMID: 27867147 Review.
-
Comparative Genomics of the Zic Family Genes.Adv Exp Med Biol. 2018;1046:3-26. doi: 10.1007/978-981-10-7311-3_1. Adv Exp Med Biol. 2018. PMID: 29442314 Review.
Cited by
-
On the dependent recognition of some long zinc finger proteins.Nucleic Acids Res. 2023 Jun 23;51(11):5364-5376. doi: 10.1093/nar/gkad207. Nucleic Acids Res. 2023. PMID: 36951113 Free PMC article.
-
An improved predictive recognition model for Cys(2)-His(2) zinc finger proteins.Nucleic Acids Res. 2014 Apr;42(8):4800-12. doi: 10.1093/nar/gku132. Epub 2014 Feb 12. Nucleic Acids Res. 2014. PMID: 24523353 Free PMC article.
-
Analysis of an artificial zinc finger epigenetic modulator: widespread binding but limited regulation.Nucleic Acids Res. 2014;42(16):10856-68. doi: 10.1093/nar/gku708. Epub 2014 Aug 13. Nucleic Acids Res. 2014. PMID: 25122745 Free PMC article.
-
What Can Domesticated Genes Tell Us about the Intron Gain in Mammals?Int J Evol Biol. 2012;2012:278981. doi: 10.1155/2012/278981. Epub 2012 May 30. Int J Evol Biol. 2012. PMID: 22693680 Free PMC article.
-
TFClass: a classification of human transcription factors and their rodent orthologs.Nucleic Acids Res. 2015 Jan;43(Database issue):D97-102. doi: 10.1093/nar/gku1064. Epub 2014 Oct 31. Nucleic Acids Res. 2015. PMID: 25361979 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

