A diversity profile of the human skin microbiota
- PMID: 18502944
- PMCID: PMC2493393
- DOI: 10.1101/gr.075549.107
A diversity profile of the human skin microbiota
Abstract
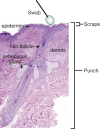
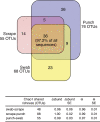
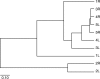
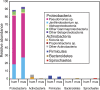
The many layers and structures of the skin serve as elaborate hosts to microbes, including a diversity of commensal and pathogenic bacteria that contribute to both human health and disease. To determine the complexity and identity of the microbes inhabiting the skin, we sequenced bacterial 16S small-subunit ribosomal RNA genes isolated from the inner elbow of five healthy human subjects. This analysis revealed 113 operational taxonomic units (OTUs; "phylotypes") at the level of 97% similarity that belong to six bacterial divisions. To survey all depths of the skin, we sampled using three methods: swab, scrape, and punch biopsy. Proteobacteria dominated the skin microbiota at all depths of sampling. Interpersonal variation is approximately equal to intrapersonal variation when considering bacterial community membership and structure. Finally, we report strong similarities in the complexity and identity of mouse and human skin microbiota. This study of healthy human skin microbiota will serve to direct future research addressing the role of skin microbiota in health and disease, and metagenomic projects addressing the complex physiological interactions between the skin and the microbes that inhabit this environment.
Figures

Similar articles
-
OTUs and ASVs Produce Comparable Taxonomic and Diversity from Shrimp Microbiota 16S Profiles Using Tailored Abundance Filters.Genes (Basel). 2021 Apr 13;12(4):564. doi: 10.3390/genes12040564. Genes (Basel). 2021. PMID: 33924545 Free PMC article.
-
Molecular analysis of the prevalent microbiota of human male and female forehead skin compared to forearm skin and the influence of make-up.J Appl Microbiol. 2011 Jun;110(6):1381-9. doi: 10.1111/j.1365-2672.2011.04991.x. Epub 2011 Mar 17. J Appl Microbiol. 2011. PMID: 21362117
-
New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome.mBio. 2019 Jul 2;10(4):e00839-19. doi: 10.1128/mBio.00839-19. mBio. 2019. PMID: 31266865 Free PMC article.
-
Chemical ecology of interactions between human skin microbiota and mosquitoes.FEMS Microbiol Ecol. 2010 Oct;74(1):1-9. doi: 10.1111/j.1574-6941.2010.00908.x. FEMS Microbiol Ecol. 2010. PMID: 20840217 Review.
-
Human Skin Is the Largest Epithelial Surface for Interaction with Microbes.J Invest Dermatol. 2017 Jun;137(6):1213-1214. doi: 10.1016/j.jid.2016.11.045. Epub 2017 Apr 8. J Invest Dermatol. 2017. PMID: 28395897 Free PMC article. Review.
Cited by
-
The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing.Int J Legal Med. 2014 Jan;128(1):193-205. doi: 10.1007/s00414-013-0872-1. Epub 2013 Jun 10. Int J Legal Med. 2014. PMID: 23749255
-
Challenging Cosmetic Innovation: The Skin Microbiota and Probiotics Protect the Skin from UV-Induced Damage.Microorganisms. 2021 Apr 27;9(5):936. doi: 10.3390/microorganisms9050936. Microorganisms. 2021. PMID: 33925587 Free PMC article. Review.
-
Experimental and analytical tools for studying the human microbiome.Nat Rev Genet. 2011 Dec 16;13(1):47-58. doi: 10.1038/nrg3129. Nat Rev Genet. 2011. PMID: 22179717 Free PMC article. Review.
-
Immunotoxicity of nanomaterials in health and disease: Current challenges and emerging approaches for identifying immune modifiers in susceptible populations.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022 Nov;14(6):e1804. doi: 10.1002/wnan.1804. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2022. PMID: 36416020 Free PMC article. Review.
-
Body Site Is a More Determinant Factor than Human Population Diversity in the Healthy Skin Microbiome.PLoS One. 2016 Apr 18;11(4):e0151990. doi: 10.1371/journal.pone.0151990. eCollection 2016. PLoS One. 2016. PMID: 27088867 Free PMC article.
References
-
- Bruggemann H., Henne A., Hoster F., Liesegang H., Wiezer A., Strittmatter A., Hujer S., Durre P., Gottschalk G. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305:671–673. - PubMed
-
- Castillo M., Martin-Orue S.M., Manzanilla E.G., Badiola I., Martin M., Gasa J. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 2006;114:165–170. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
