Proteasome subunit Rpn13 is a novel ubiquitin receptor
- PMID: 18497817
- PMCID: PMC2839886
- DOI: 10.1038/nature06926
Proteasome subunit Rpn13 is a novel ubiquitin receptor
Abstract
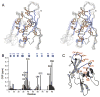
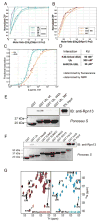
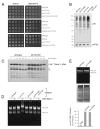
Proteasomal receptors that recognize ubiquitin chains attached to substrates are key mediators of selective protein degradation in eukaryotes. Here we report the identification of a new ubiquitin receptor, Rpn13/ARM1, a known component of the proteasome. Rpn13 binds ubiquitin through a conserved amino-terminal region termed the pleckstrin-like receptor for ubiquitin (Pru) domain, which binds K48-linked diubiquitin with an affinity of approximately 90 nM. Like proteasomal ubiquitin receptor Rpn10/S5a, Rpn13 also binds ubiquitin-like (UBL) domains of UBL-ubiquitin-associated (UBA) proteins. In yeast, a synthetic phenotype results when specific mutations of the ubiquitin binding sites of Rpn10 and Rpn13 are combined, indicating functional linkage between these ubiquitin receptors. Because Rpn13 is also the proteasomal receptor for Uch37, a deubiquitinating enzyme, our findings suggest a coupling of chain recognition and disassembly at the proteasome.
Figures

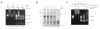

Comment in
-
Cell biology: two hands for degradation.Nature. 2008 May 22;453(7194):460-1. doi: 10.1038/453460a. Nature. 2008. PMID: 18497808 No abstract available.
Similar articles
-
Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.Nature. 2008 May 22;453(7194):548-52. doi: 10.1038/nature06924. Nature. 2008. PMID: 18497827 Free PMC article.
-
Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.Mol Cell. 2009 Aug 14;35(3):280-90. doi: 10.1016/j.molcel.2009.06.010. Mol Cell. 2009. PMID: 19683493 Free PMC article.
-
Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.J Biol Chem. 2017 Jun 9;292(23):9493-9504. doi: 10.1074/jbc.M117.785287. Epub 2017 Apr 25. J Biol Chem. 2017. PMID: 28442575 Free PMC article.
-
Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation.Cell Mol Life Sci. 2009 Sep;66(17):2819-33. doi: 10.1007/s00018-009-0048-9. Epub 2009 May 26. Cell Mol Life Sci. 2009. PMID: 19468686 Free PMC article. Review.
-
Proteins directly interacting with mammalian 20S proteasomal subunits and ubiquitin-independent proteasomal degradation.Biomolecules. 2014 Dec 19;4(4):1140-54. doi: 10.3390/biom4041140. Biomolecules. 2014. PMID: 25534281 Free PMC article. Review.
Cited by
-
Functions of the 19S complex in proteasomal degradation.Trends Biochem Sci. 2013 Feb;38(2):103-10. doi: 10.1016/j.tibs.2012.11.009. Epub 2013 Jan 2. Trends Biochem Sci. 2013. PMID: 23290100 Free PMC article. Review.
-
Co-Chaperones in Targeting and Delivery of Misfolded Proteins to the 26S Proteasome.Biomolecules. 2020 Aug 4;10(8):1141. doi: 10.3390/biom10081141. Biomolecules. 2020. PMID: 32759676 Free PMC article. Review.
-
Impact of Losing hRpn13 Pru or UCHL5 on Proteasome Clearance of Ubiquitinated Proteins and RA190 Cytotoxicity.Mol Cell Biol. 2020 Aug 28;40(18):e00122-20. doi: 10.1128/MCB.00122-20. Print 2020 Aug 28. Mol Cell Biol. 2020. PMID: 32631902 Free PMC article.
-
Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis.PLoS Genet. 2015 Jul 29;11(7):e1005401. doi: 10.1371/journal.pgen.1005401. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26222436 Free PMC article.
-
hRpn13 shapes the proteome and transcriptome through epigenetic factors HDAC8, PADI4, and transcription factor NF-κB p50.Mol Cell. 2024 Feb 1;84(3):522-537.e8. doi: 10.1016/j.molcel.2023.11.035. Epub 2023 Dec 26. Mol Cell. 2024. PMID: 38151017
References
-
- Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–68. - PubMed
-
- Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–61. - PubMed
-
- Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–22. - PubMed
-
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. - PubMed
-
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–9. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

