Modification of Drosophila p53 by SUMO modulates its transactivation and pro-apoptotic functions
- PMID: 18492669
- PMCID: PMC3258953
- DOI: 10.1074/jbc.M710186200
Modification of Drosophila p53 by SUMO modulates its transactivation and pro-apoptotic functions
Abstract
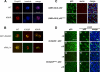
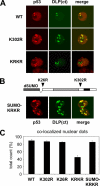
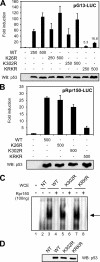
Conjugation to SUMO is a reversible post-translational modification that regulates several transcription factors involved in cell proliferation, differentiation, and disease. The p53 tumor suppressor can be modified by SUMO-1 in mammalian cells, but the functional consequences of this modification are unclear. Here, we demonstrate that the Drosophila homolog of human p53 can be efficiently sumoylated in insect cells. We identify two lysine residues involved in SUMO attachment, one at the C terminus, between the DNA binding and oligomerization domains, and one at the N terminus of the protein. We find that sumoylation helps recruit Drosophila p53 to nuclear dot-like structures that can be marked by human PML and the Drosophila homologue of Daxx. We demonstrate that mutation of both sumoylation sites dramatically reduces the transcriptional activity of p53 and its ability to induce apoptosis in transgenic flies, providing in vivo evidence that sumoylation is critical for Drosophila p53 function.
Figures

Similar articles
-
Identification of two independent SUMO-interacting motifs in Daxx: evolutionary conservation from Drosophila to humans and their biochemical functions.Cell Cycle. 2009 Jan 1;8(1):76-87. doi: 10.4161/cc.8.1.7493. Cell Cycle. 2009. PMID: 19106612
-
Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors.Mol Cell. 2006 Nov 3;24(3):341-54. doi: 10.1016/j.molcel.2006.10.019. Mol Cell. 2006. PMID: 17081986
-
SUMO-mediated inhibition of glucocorticoid receptor synergistic activity depends on stable assembly at the promoter but not on DAXX.Mol Endocrinol. 2008 Sep;22(9):2061-75. doi: 10.1210/me.2007-0581. Epub 2008 Jun 18. Mol Endocrinol. 2008. PMID: 18562626 Free PMC article.
-
Daxx mediates SUMO-dependent transcriptional control and subnuclear compartmentalization.Biochem Soc Trans. 2007 Dec;35(Pt 6):1397-400. doi: 10.1042/BST0351397. Biochem Soc Trans. 2007. PMID: 18031230 Review.
-
Regulation of p53 family members by the ubiquitin-like SUMO system.DNA Repair (Amst). 2009 Apr 5;8(4):491-8. doi: 10.1016/j.dnarep.2009.01.002. Epub 2009 Feb 12. DNA Repair (Amst). 2009. PMID: 19213614 Review.
Cited by
-
Drosophila p53 isoforms have overlapping and distinct functions in germline genome integrity and oocyte quality control.Elife. 2022 Jan 13;11:e61389. doi: 10.7554/eLife.61389. Elife. 2022. PMID: 35023826 Free PMC article.
-
Classification of intrinsically disordered regions and proteins.Chem Rev. 2014 Jul 9;114(13):6589-631. doi: 10.1021/cr400525m. Epub 2014 Apr 29. Chem Rev. 2014. PMID: 24773235 Free PMC article. Review. No abstract available.
-
SUMO interaction motifs in Sizn1 are required for promyelocytic leukemia protein nuclear body localization and for transcriptional activation.J Biol Chem. 2009 Jul 17;284(29):19592-600. doi: 10.1074/jbc.M109.010181. Epub 2009 May 5. J Biol Chem. 2009. PMID: 19416967 Free PMC article.
-
Sumo-2/3-ylation following in vitro modeled ischemia is reduced in delayed ischemic tolerance.Brain Res. 2009 May 26;1272:71-80. doi: 10.1016/j.brainres.2009.03.034. Epub 2009 Mar 28. Brain Res. 2009. PMID: 19332039 Free PMC article.
-
In vivo analysis of a fluorescent SUMO fusion in transgenic Drosophila.Fly (Austin). 2014;8(2):108-12. doi: 10.4161/fly.28312. Fly (Austin). 2014. PMID: 25483255 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

