Similar active genes cluster in specialized transcription factories
- PMID: 18490511
- PMCID: PMC2386102
- DOI: 10.1083/jcb.200710053
Similar active genes cluster in specialized transcription factories
Abstract
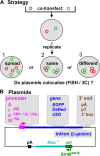
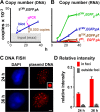
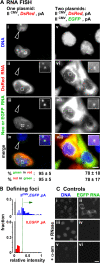
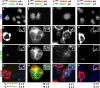
How transcription affects the way specific genes are arranged within the nucleus remains to be fully understood. We examine here whether transcription occurs in discrete sites (factories) containing the required machinery and whether these sites specialize in transcribing different genes. We cotransfected plasmids encoding a common origin of replication but different transcription units into cells, where they are assembled into minichromosomes that the cellular machinery replicates and transcribes. In cells containing thousands of minichromosomes, we found (using fluorescence in situ hybridization) active templates concentrated in only a few factories that transcribe particular units depending on the promoter type and the presence of an intron. Close proximity between similar transcription units, whether on two different minichromosomes or on host chromosomes and minichromosomes, is confirmed using chromosome conformation capture. We conclude that factories specialize in producing a particular type of transcript depending on promoter type and whether or not the gene contains an intron.
Figures

Similar articles
-
RNA polymerase I (Pol I) passage through nucleosomes depends on Pol I subunits binding its lobe structure.J Biol Chem. 2020 Apr 10;295(15):4782-4795. doi: 10.1074/jbc.RA119.011827. Epub 2020 Feb 14. J Biol Chem. 2020. PMID: 32060094 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Hijacks RNA Polymerase II To Create a Viral Transcriptional Factory.J Virol. 2017 May 12;91(11):e02491-16. doi: 10.1128/JVI.02491-16. Print 2017 Jun 1. J Virol. 2017. PMID: 28331082 Free PMC article.
-
RNA polymerase III promoter elements enhance transcription of RNA polymerase II genes.Nucleic Acids Res. 1988 Feb 25;16(4):1285-93. doi: 10.1093/nar/16.4.1285. Nucleic Acids Res. 1988. PMID: 2831496 Free PMC article.
-
Nuclear organization of RNA polymerase II transcription.Biochem Cell Biol. 2013 Feb;91(1):22-30. doi: 10.1139/bcb-2012-0059. Epub 2013 Feb 5. Biochem Cell Biol. 2013. PMID: 23442138 Review.
-
The role of specialized transcription factories in chromosome pairing.Biochim Biophys Acta. 2008 Nov;1783(11):2155-60. doi: 10.1016/j.bbamcr.2008.07.013. Epub 2008 Jul 28. Biochim Biophys Acta. 2008. PMID: 18706455 Review.
Cited by
-
Understanding the regulatory and transcriptional complexity of the genome through structure.Genome Res. 2013 Jul;23(7):1081-8. doi: 10.1101/gr.156612.113. Genome Res. 2013. PMID: 23817049 Free PMC article. Review.
-
Arid5a cooperates with Sox9 to stimulate chondrocyte-specific transcription.Mol Biol Cell. 2011 Apr 15;22(8):1300-11. doi: 10.1091/mbc.E10-07-0566. Epub 2011 Feb 23. Mol Biol Cell. 2011. PMID: 21346191 Free PMC article.
-
Location and dynamics of an active promoter in Escherichia coli K-12.Biochem J. 2012 Jan 1;441(1):481-5. doi: 10.1042/BJ20111258. Biochem J. 2012. PMID: 21936772 Free PMC article.
-
Organization of the amplified type I interferon gene cluster and associated chromosome regions in the interphase nucleus of human osteosarcoma cells.Chromosome Res. 2009;17(3):305-19. doi: 10.1007/s10577-009-9023-4. Epub 2009 Mar 13. Chromosome Res. 2009. PMID: 19283497 Free PMC article.
-
The pluripotent genome in three dimensions is shaped around pluripotency factors.Nature. 2013 Sep 12;501(7466):227-31. doi: 10.1038/nature12420. Epub 2013 Jul 24. Nature. 2013. PMID: 23883933
References
-
- Barski, A., S. Cuddapah, K. Cui, T.Y. Roh, D.E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell. 129:823–837. - PubMed
-
- Bartlett, J., J. Blagojevic, D. Carter, C. Eskiw, M. Fromaget, C. Job, M. Shamsher, I.F. Trindade, M. Xu, and P.R. Cook. 2006. Specialized transcription factories. Biochem. Soc. Symp. 73:67–75. - PubMed
-
- Binnie, A., P. Castelo-Branco, J. Monks, and N.J. Proudfoot. 2006. Homologous gene sequences mediate transcription-domain formation. J. Cell Sci. 119:3876–3887. - PubMed
-
- Boyd, D.C., P.C. Turner, N.J. Watkins, T. Gerster, and S. Murphy. 1995. Functional redundancy of promoter elements ensures efficient transcription of the human 7SK gene in vivo. J. Mol. Biol. 253:677–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

