QUMA: quantification tool for methylation analysis
- PMID: 18487274
- PMCID: PMC2447804
- DOI: 10.1093/nar/gkn294
QUMA: quantification tool for methylation analysis
Abstract
Bisulfite sequencing, a standard method for DNA methylation profile analysis, is widely used in basic and clinical studies. This method is limited, however, by the time-consuming data analysis processes required to obtain accurate DNA methylation profiles from the raw sequence output of the DNA sequencer, and by the fact that quality checking of the results can be influenced by a researcher's bias. We have developed an interactive and easy-to-use web-based tool, QUMA (quantification tool for methylation analysis), for the bisulfite sequencing analysis of CpG methylation. QUMA includes most of the data-processing functions necessary for the analysis of bisulfite sequences. It also provides a platform for consistent quality control of the analysis. The QUMA web server is available at http://quma.cdb.riken.jp/.
Figures


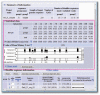
Similar articles
-
BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing.Bioinformatics. 2005 Nov 1;21(21):4067-8. doi: 10.1093/bioinformatics/bti652. Epub 2005 Sep 1. Bioinformatics. 2005. PMID: 16141249
-
ampliMethProfiler: a pipeline for the analysis of CpG methylation profiles of targeted deep bisulfite sequenced amplicons.BMC Bioinformatics. 2016 Nov 25;17(1):484. doi: 10.1186/s12859-016-1380-3. BMC Bioinformatics. 2016. PMID: 27884103 Free PMC article.
-
BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences.BMC Bioinformatics. 2010 May 6;11:230. doi: 10.1186/1471-2105-11-230. BMC Bioinformatics. 2010. PMID: 20459626 Free PMC article.
-
BiSearch: ePCR tool for native or bisulfite-treated genomic template.Methods Mol Biol. 2007;402:385-402. doi: 10.1007/978-1-59745-528-2_20. Methods Mol Biol. 2007. PMID: 17951807 Review.
-
Methods for CpG Methylation Array Profiling Via Bisulfite Conversion.Methods Mol Biol. 2018;1706:233-254. doi: 10.1007/978-1-4939-7471-9_13. Methods Mol Biol. 2018. PMID: 29423802 Free PMC article. Review.
Cited by
-
Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method.Nucleic Acids Res. 2013 Apr;41(7):e84. doi: 10.1093/nar/gkt091. Epub 2013 Feb 13. Nucleic Acids Res. 2013. PMID: 23408859 Free PMC article.
-
Characterization of the regulatory 5'-flanking region of bovine mucin 2 (MUC2) gene.Mol Cell Biochem. 2021 Jul;476(7):2847-2856. doi: 10.1007/s11010-021-04133-1. Epub 2021 Mar 17. Mol Cell Biochem. 2021. PMID: 33730299
-
Regulation of DNA methylation dictates Cd4 expression during the development of helper and cytotoxic T cell lineages.Nat Immunol. 2015 Jul;16(7):746-54. doi: 10.1038/ni.3198. Epub 2015 Jun 1. Nat Immunol. 2015. PMID: 26030024 Free PMC article.
-
TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer's disease hippocampus.Clin Epigenetics. 2016 Apr 5;8:37. doi: 10.1186/s13148-016-0202-9. eCollection 2016. Clin Epigenetics. 2016. PMID: 27051467 Free PMC article.
-
CDCA7-associated global aberrant DNA hypomethylation translates to localized, tissue-specific transcriptional responses.Sci Adv. 2024 Feb 9;10(6):eadk3384. doi: 10.1126/sciadv.adk3384. Epub 2024 Feb 9. Sci Adv. 2024. PMID: 38335290 Free PMC article.
References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr. Opin. Cell. Biol. 2007;19:273–280. - PubMed
-
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

