Protein folding: independent unrelated pathways or predetermined pathway with optional errors
- PMID: 18480257
- PMCID: PMC2438224
- DOI: 10.1073/pnas.0801864105
Protein folding: independent unrelated pathways or predetermined pathway with optional errors
Abstract
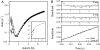
The observation of heterogeneous protein folding kinetics has been widely interpreted in terms of multiple independent unrelated pathways (IUP model), both experimentally and in theoretical calculations. However, direct structural information on folding intermediates and their properties now indicates that all of a protein population folds through essentially the same stepwise pathway, determined by cooperative native-like foldon units and the way that the foldons fit together in the native protein. It is essential to decide between these fundamentally different folding mechanisms. This article shows, contrary to previous supposition, that the heterogeneous folding kinetics observed for the staphylococcal nuclease protein (SNase) does not require alternative parallel pathways. SNase folding kinetics can be fit equally well by a single predetermined pathway that allows for optional misfolding errors, which are known to occur ubiquitously in protein folding. Structural, kinetic, and thermodynamic information for the folding intermediates and pathways of many proteins is consistent with the predetermined pathway-optional error (PPOE) model but contrary to the properties implied in IUP models.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
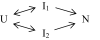

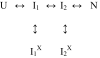
Similar articles
-
Protein folding and misfolding: mechanism and principles.Q Rev Biophys. 2007 Nov;40(4):287-326. doi: 10.1017/S0033583508004654. Epub 2008 Apr 14. Q Rev Biophys. 2007. PMID: 18405419 Free PMC article. Review.
-
Least activation path for protein folding: investigation of staphylococcal nuclease folding by stopped-flow circular dichroism.Proc Natl Acad Sci U S A. 1996 Mar 19;93(6):2539-44. doi: 10.1073/pnas.93.6.2539. Proc Natl Acad Sci U S A. 1996. PMID: 8637910 Free PMC article.
-
Fluorescence energy transfer indicates similar transient and equilibrium intermediates in staphylococcal nuclease folding.J Mol Biol. 2000 Jun 16;299(4):1133-46. doi: 10.1006/jmbi.2000.3804. J Mol Biol. 2000. PMID: 10843864
-
Kinetic evidence for folding and unfolding intermediates in staphylococcal nuclease.Biochemistry. 1997 May 13;36(19):5795-805. doi: 10.1021/bi9700476. Biochemistry. 1997. PMID: 9153420
-
The nature of protein folding pathways.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):15873-80. doi: 10.1073/pnas.1411798111. Epub 2014 Oct 17. Proc Natl Acad Sci U S A. 2014. PMID: 25326421 Free PMC article. Review.
Cited by
-
Stability of uniformly labeled (13C and 15N) cytochrome c and its L94G mutant.Sci Rep. 2021 Mar 24;11(1):6804. doi: 10.1038/s41598-021-86332-w. Sci Rep. 2021. PMID: 33762670 Free PMC article.
-
How cooperative are protein folding and unfolding transitions?Protein Sci. 2016 Nov;25(11):1924-1941. doi: 10.1002/pro.3015. Epub 2016 Sep 13. Protein Sci. 2016. PMID: 27522064 Free PMC article. Review.
-
Quantitative assessment of protein structural models by comparison of H/D exchange MS data with exchange behavior accurately predicted by DXCOREX.J Am Soc Mass Spectrom. 2012 Jan;23(1):43-56. doi: 10.1007/s13361-011-0267-9. Epub 2011 Oct 20. J Am Soc Mass Spectrom. 2012. PMID: 22012689 Free PMC article.
-
Intra-molecular chaperone: the role of the N-terminal in conformational selection and kinetic control.Phys Biol. 2009 Feb 4;6(1):013001. doi: 10.1088/1478-3975/6/1/013001. Phys Biol. 2009. PMID: 19193974 Free PMC article.
-
How main-chains of proteins explore the free-energy landscape in native states.Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19708-13. doi: 10.1073/pnas.0810679105. Epub 2008 Dec 10. Proc Natl Acad Sci U S A. 2008. PMID: 19073932 Free PMC article.
References
-
- Levinthal C. Are there pathways for protein folding? J Chim Phys. 1968;65:44–45.
-
- Kim PS, Baldwin RL. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. - PubMed
-
- Wolynes PG, Onuchic JN, Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

