A transcriptional enhancer from the coding region of ADAMTS5
- PMID: 18478108
- PMCID: PMC2364661
- DOI: 10.1371/journal.pone.0002184
A transcriptional enhancer from the coding region of ADAMTS5
Abstract
Background: The revelation that the human genome encodes only approximately 25,000 genes and thus cannot account for phenotypic complexity has been one of the biggest surprises in the post-genomic era. However, accumulating evidence suggests that transcriptional regulation may be in large part responsible for this observed mammalian complexity. Consequently, there has been a strong drive to locate cis-regulatory regions in mammalian genomes in order to understand the unifying principles governing these regions, including their genomic distribution. Although a number of systematic approaches have been developed, these all discount coding sequence.
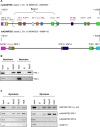
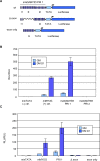
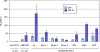
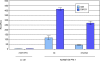
Methodology/principal findings: Using the computational tool PRI (Pattern-defined Regulatory Islands), which does not mask coding sequence, we identified a regulatory region associated with the gene ADAMTS5 that encompasses the entirety of the essential coding exon 2. We demonstrate through a combination of chromatin immunoprecipitation and reporter gene studies that this region can not only bind the myogenic transcription factors MYOD and myogenin and the E-protein HEB but can also function as a very strong myogenic transcriptional enhancer.
Conclusions/significance: Thus, we report the identification and detailed characterization of an exonic enhancer. Ultimately, this leads to the interesting question of why evolution would be so parsimonious in the functional assignment of sequence.
Conflict of interest statement
Figures
Similar articles
-
Identifying pattern-defined regulatory islands in mammalian genomes.Proc Natl Acad Sci U S A. 2007 Jun 12;104(24):10116-21. doi: 10.1073/pnas.0704028104. Epub 2007 May 29. Proc Natl Acad Sci U S A. 2007. PMID: 17535887 Free PMC article.
-
Exonic remnants of whole-genome duplication reveal cis-regulatory function of coding exons.Nucleic Acids Res. 2010 Mar;38(4):1071-85. doi: 10.1093/nar/gkp1124. Epub 2009 Dec 6. Nucleic Acids Res. 2010. PMID: 19969543 Free PMC article.
-
A novel myogenic regulatory circuit controls slow/cardiac troponin C gene transcription in skeletal muscle.Mol Cell Biol. 1994 Mar;14(3):1870-85. doi: 10.1128/mcb.14.3.1870-1885.1994. Mol Cell Biol. 1994. PMID: 8114720 Free PMC article.
-
Activation of muscle-specific transcription by myogenic helix-loop-helix proteins.Symp Soc Exp Biol. 1992;46:331-41. Symp Soc Exp Biol. 1992. PMID: 1341046 Review.
-
Noncoding transcription at enhancers: general principles and functional models.Annu Rev Genet. 2012;46:1-19. doi: 10.1146/annurev-genet-110711-155459. Epub 2012 Aug 16. Annu Rev Genet. 2012. PMID: 22905871 Review.
Cited by
-
IP(3)-dependent, post-tetanic calcium transients induced by electrostimulation of adult skeletal muscle fibers.J Gen Physiol. 2010 Oct;136(4):455-67. doi: 10.1085/jgp.200910397. Epub 2010 Sep 13. J Gen Physiol. 2010. PMID: 20837675 Free PMC article.
-
DNA shape, genetic codes, and evolution.Curr Opin Struct Biol. 2011 Jun;21(3):342-7. doi: 10.1016/j.sbi.2011.03.002. Epub 2011 Mar 23. Curr Opin Struct Biol. 2011. PMID: 21439813 Free PMC article. Review.
-
Transcriptional coupling of synaptic transmission and energy metabolism: role of nuclear respiratory factor 1 in co-regulating neuronal nitric oxide synthase and cytochrome c oxidase genes in neurons.Biochim Biophys Acta. 2009 Oct;1793(10):1604-13. doi: 10.1016/j.bbamcr.2009.07.001. Epub 2009 Jul 14. Biochim Biophys Acta. 2009. PMID: 19615412 Free PMC article.
-
Identification of a Cis-acting regulatory polymorphism in a Eucalypt COBRA-like gene affecting cellulose content.Genetics. 2009 Nov;183(3):1153-64. doi: 10.1534/genetics.109.106591. Epub 2009 Sep 7. Genetics. 2009. PMID: 19737751 Free PMC article.
-
Transcriptional induction of ADAMTS5 protein by nuclear factor-κB (NF-κB) family member RelA/p65 in chondrocytes during osteoarthritis development.J Biol Chem. 2013 Oct 4;288(40):28620-9. doi: 10.1074/jbc.M113.452169. Epub 2013 Aug 20. J Biol Chem. 2013. PMID: 23963448 Free PMC article.
References
-
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. - PubMed
-
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
-
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. - PubMed
-
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

