Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding
- PMID: 18474528
- PMCID: PMC2441811
- DOI: 10.1093/nar/gkn249
Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding
Abstract
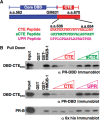
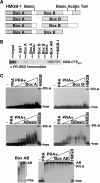
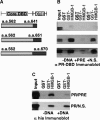
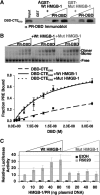
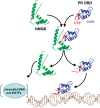
The DNA-binding domain (DBD) of progesterone receptor (PR) is bipartite containing a zinc module core that interacts with progesterone response elements (PRE), and a short flexible carboxyl terminal extension (CTE) that interacts with the minor groove flanking the PRE. The chromosomal high-mobility group B proteins (HMGB), defined as DNA architectural proteins capable of bending DNA, also function as auxiliary factors that increase the DNA-binding affinity of PR and other steroid receptors by mechanisms that are not well defined. Here we show that the CTE of PR contains a specific binding site for HMGB that is required for stimulation of PR-PRE binding, whereas the DNA architectural properties of HMGB are dispensable. Specific PRE DNA inhibited HMGB binding to the CTE, indicating that DNA and HMGB-CTE interactions are mutually exclusive. Exogenous CTE peptide increased PR-binding affinity for PRE as did deletion of the CTE. In a PR-binding site selection assay, A/T sequences flanking the PRE were enriched by HMGB, indicating that PR DNA-binding specificity is also altered by HMGB. We conclude that a transient HMGB-CTE interaction alters a repressive conformation of the flexible CTE enabling it to bind to preferred sequences flanking the PRE.
Figures


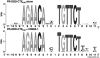

Similar articles
-
Structural and functional analysis of domains of the progesterone receptor.Mol Cell Endocrinol. 2012 Jan 30;348(2):418-29. doi: 10.1016/j.mce.2011.07.017. Epub 2011 Jul 22. Mol Cell Endocrinol. 2012. PMID: 21803119 Free PMC article. Review.
-
The role of the C-terminal extension (CTE) of the estrogen receptor alpha and beta DNA binding domain in DNA binding and interaction with HMGB.J Biol Chem. 2004 Apr 9;279(15):14763-71. doi: 10.1074/jbc.M313335200. Epub 2004 Jan 21. J Biol Chem. 2004. PMID: 14739282
-
A progesterone receptor co-activator (JDP2) mediates activity through interaction with residues in the carboxyl-terminal extension of the DNA binding domain.J Biol Chem. 2009 Sep 4;284(36):24415-24. doi: 10.1074/jbc.M109.003244. Epub 2009 Jun 24. J Biol Chem. 2009. PMID: 19553667 Free PMC article.
-
Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements.Mol Endocrinol. 2006 Dec;20(12):3042-52. doi: 10.1210/me.2005-0511. Epub 2006 Aug 24. Mol Endocrinol. 2006. PMID: 16931575 Free PMC article.
-
Chromatin-associated HMGA and HMGB proteins: versatile co-regulators of DNA-dependent processes.Plant Mol Biol. 2003 Oct;53(3):281-95. doi: 10.1023/b:plan.0000007002.99408.ba. Plant Mol Biol. 2003. PMID: 14750519 Review.
Cited by
-
Nucleosome dynamics: HMGB1 relaxes canonical nucleosome structure to facilitate estrogen receptor binding.Nucleic Acids Res. 2012 Nov 1;40(20):10161-71. doi: 10.1093/nar/gks815. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941653 Free PMC article.
-
Transcriptional regulation: it takes a village.Mol Cell. 2008 Sep 5;31(5):622-9. doi: 10.1016/j.molcel.2008.08.013. Mol Cell. 2008. PMID: 18775322 Free PMC article. Review.
-
The high mobility group box: the ultimate utility player of a cell.Trends Biochem Sci. 2012 Dec;37(12):553-62. doi: 10.1016/j.tibs.2012.09.003. Epub 2012 Nov 13. Trends Biochem Sci. 2012. PMID: 23153957 Free PMC article. Review.
-
Structural and functional analysis of domains of the progesterone receptor.Mol Cell Endocrinol. 2012 Jan 30;348(2):418-29. doi: 10.1016/j.mce.2011.07.017. Epub 2011 Jul 22. Mol Cell Endocrinol. 2012. PMID: 21803119 Free PMC article. Review.
-
The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention.Mol Endocrinol. 2010 Nov;24(11):2126-38. doi: 10.1210/me.2010-0170. Epub 2010 Sep 22. Mol Endocrinol. 2010. PMID: 20861224 Free PMC article.
References
-
- Beato M, Klug J. Steroid hormone receptors: an update. Hum. Reprod. Update. 2000;6:225–236. - PubMed
-
- Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 2001;26:384–390. - PubMed
-
- Gearhart MD, Holmbeck SM, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J. Mol. Biol. 2003;327:819–832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

