Are DNA transcription factor proteins maxwellian demons?
- PMID: 18456820
- PMCID: PMC2479577
- DOI: 10.1529/biophysj.108.129825
Are DNA transcription factor proteins maxwellian demons?
Abstract
Transcription factor (TF) proteins rapidly locate unique target sites on long genomic DNA molecules--and bind to them--during gene regulation. The search mechanism is known to involve a combination of three-dimensional diffusion through the bulk of the cell and one-dimensional sliding diffusion along the DNA. It is believed that the surprisingly high target binding rates of TF proteins relies on conformational fluctuations of the protein between a mobile state that is insensitive to the DNA sequence and an immobile state that is sequence-sensitive. Since TFs are not able to consume free energy during their search to obtain DNA sequence information, the Second Law of Thermodynamics must impose a strict limit on the efficiency of passive search mechanisms. In this article, we use a simple model for the protein conformational fluctuations to obtain the shortest binding time consistent with thermodynamics. The binding time is minimized if the spectrum of conformational fluctuations that take place during the search is impedance-matched to the large-scale conformational change that takes place at the target site. For parameter values appropriate for bacterial TF, this minimum binding time is within an order-of-magnitude of a limiting binding time corresponding to an idealized protein with instant target recognition. Numerical estimates suggest that typical bacteria operate in this regime of optimized conformational fluctuations.
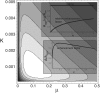
Figures
Similar articles
-
Facilitated diffusion on confined DNA.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Feb;85(2 Pt 1):021919. doi: 10.1103/PhysRevE.85.021919. Epub 2012 Feb 22. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 22463256
-
Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential.Biophys J. 2004 Dec;87(6):4021-35. doi: 10.1529/biophysj.104.050765. Epub 2004 Oct 1. Biophys J. 2004. PMID: 15465864 Free PMC article.
-
Generalized theory of site-specific DNA-protein interactions.Phys Rev E Stat Nonlin Soft Matter Phys. 2007 Jul;76(1 Pt 1):011901. doi: 10.1103/PhysRevE.76.011901. Epub 2007 Jul 2. Phys Rev E Stat Nonlin Soft Matter Phys. 2007. PMID: 17677488
-
How motif environment influences transcription factor search dynamics: Finding a needle in a haystack.Bioessays. 2016 Jul;38(7):605-12. doi: 10.1002/bies.201600005. Epub 2016 May 19. Bioessays. 2016. PMID: 27192961 Free PMC article. Review.
-
Towards an understanding of protein-DNA recognition.Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):501-9. doi: 10.1098/rstb.1996.0048. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735272 Review.
Cited by
-
On the Possibility of Facilitated Diffusion of Dendrimers Along DNA.J Phys Chem B. 2015 Jun 11;119(23):6894-904. doi: 10.1021/acs.jpcb.5b02090. Epub 2015 Jun 2. J Phys Chem B. 2015. PMID: 25989627 Free PMC article.
-
Positive and negative design for nonconsensus protein-DNA binding affinity in the vicinity of functional binding sites.Biophys J. 2013 Oct 1;105(7):1653-60. doi: 10.1016/j.bpj.2013.08.033. Biophys J. 2013. PMID: 24094406 Free PMC article.
-
Tethered-hopping model for protein-DNA binding and unbinding based on Sox2-Oct1-Hoxb1 ternary complex simulations.Biophys J. 2010 Apr 7;98(7):1285-93. doi: 10.1016/j.bpj.2009.12.4274. Biophys J. 2010. PMID: 20371328 Free PMC article.
-
Pathways for virus assembly around nucleic acids.J Mol Biol. 2014 Sep 9;426(18):3148-3165. doi: 10.1016/j.jmb.2014.07.004. Epub 2014 Jul 16. J Mol Biol. 2014. PMID: 25036288 Free PMC article.
-
Proteins searching for their target on DNA by one-dimensional diffusion: overcoming the "speed-stability" paradox.J Biol Phys. 2013 Jun;39(3):565-86. doi: 10.1007/s10867-013-9310-3. Epub 2013 Apr 5. J Biol Phys. 2013. PMID: 23860925 Free PMC article.
References
-
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 1994. Molecular Biology of the Cell. Garland, New York.
-
- Adam, G., and M. Delbrück. 1968. Structural Chemistry and Molecular Biology. A. Rich and N. Davidson, editors. Freeman, New York.
-
- Riggs, A. D., S. Bourgeois, and M. Cohn. 1970. The lac repressor-operator interaction: III. kinetic studies. J. Mol. Biol. 53:401–417. - PubMed
-
- Richter, P. H., and M. Eigen. 1974. Diffusion controlled reaction rates in spheroidal geometry: application to repressor-operator association and membrane bound enzymes. Biophys. Chem. 2:255–263. - PubMed
-
- Berg, O. G., R. B. Winter, and P. H. von Hippel. 1981. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 20:6929–6948. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

