Two strategies for gene regulation by promoter nucleosomes
- PMID: 18448704
- PMCID: PMC2493397
- DOI: 10.1101/gr.076059.108
Two strategies for gene regulation by promoter nucleosomes
Abstract
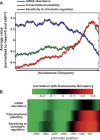
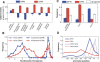
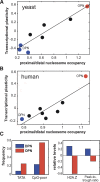
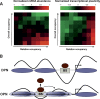
Chromatin structure is central for the regulation of gene expression, but its genome-wide organization is only beginning to be understood. Here, we examine the connection between patterns of nucleosome occupancy and the capacity to modulate gene expression upon changing conditions, i.e., transcriptional plasticity. By analyzing genome-wide data of nucleosome positioning in yeast, we find that the presence of nucleosomes close to the transcription start site is associated with high transcriptional plasticity, while nucleosomes at more distant upstream positions are negatively correlated with transcriptional plasticity. Based on this, we identify two typical promoter structures associated with low or high plasticity, respectively. The first class is characterized by a relatively large nucleosome-free region close to the start site coupled with well-positioned nucleosomes further upstream, whereas the second class displays a more evenly distributed and dynamic nucleosome positioning, with high occupancy close to the start site. The two classes are further distinguished by multiple promoter features, including histone turnover, binding site locations, H2A.Z occupancy, expression noise, and expression diversity. Analysis of nucleosome positioning in human promoters reproduces the main observations. Our results suggest two distinct strategies for gene regulation by chromatin, which are selectively employed by different genes.
Figures

Similar articles
-
New insights into two distinct nucleosome distributions: comparison of cross-platform positioning datasets in the yeast genome.BMC Genomics. 2010 Jan 15;11:33. doi: 10.1186/1471-2164-11-33. BMC Genomics. 2010. PMID: 20078849 Free PMC article.
-
Histone variant selectivity at the transcription start site: H2A.Z or H2A.Lap1.Nucleus. 2013 Nov-Dec;4(6):431-8. doi: 10.4161/nucl.26862. Epub 2013 Nov 8. Nucleus. 2013. PMID: 24213378 Free PMC article.
-
Two distinct modes of nucleosome modulation associated with different degrees of dependence of nucleosome positioning on the underlying DNA sequence.BMC Genomics. 2009 Jan 10;10:15. doi: 10.1186/1471-2164-10-15. BMC Genomics. 2009. PMID: 19134214 Free PMC article.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
-
The specificity of H2A.Z occupancy in the yeast genome and its relationship to transcription.Curr Genet. 2020 Oct;66(5):939-944. doi: 10.1007/s00294-020-01087-7. Epub 2020 Jun 14. Curr Genet. 2020. PMID: 32537667 Review.
Cited by
-
Chromatin mediation of a transcriptional memory effect in yeast.G3 (Bethesda). 2015 Mar 5;5(5):829-38. doi: 10.1534/g3.115.017418. G3 (Bethesda). 2015. PMID: 25748434 Free PMC article.
-
Orthogonal control of expression mean and variance by epigenetic features at different genomic loci.Mol Syst Biol. 2015 May 5;11(5):806. doi: 10.15252/msb.20145704. Mol Syst Biol. 2015. PMID: 25943345 Free PMC article.
-
HIV promoter integration site primarily modulates transcriptional burst size rather than frequency.PLoS Comput Biol. 2010 Sep 30;6(9):e1000952. doi: 10.1371/journal.pcbi.1000952. PLoS Comput Biol. 2010. PMID: 20941390 Free PMC article.
-
Robustness of transcriptional regulatory program influences gene expression variability.BMC Genomics. 2009 Dec 2;10:573. doi: 10.1186/1471-2164-10-573. BMC Genomics. 2009. PMID: 19954511 Free PMC article.
-
YPA: an integrated repository of promoter features in Saccharomyces cerevisiae.Nucleic Acids Res. 2011 Jan;39(Database issue):D647-52. doi: 10.1093/nar/gkq1086. Epub 2010 Nov 2. Nucleic Acids Res. 2011. PMID: 21045055 Free PMC article.
References
-
- Albert I., Mavrich T.N., Tomsho L.P., Qi J., Zanton S.J., Schuster S.C., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Basehoar A.D., Zanton S.J., Pugh B.F. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. - PubMed
-
- Batada N.N., Hurst L.D. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat. Genet. 2007;39:945–949. - PubMed
-
- Beyer A., Hollunder J., Nasheuer H.P., Wilhelm T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics. 2004;3:1083–1092. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
