Comparison of G-to-A mutation frequencies induced by APOBEC3 proteins in H9 cells and peripheral blood mononuclear cells in the context of impaired processivities of drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants
- PMID: 18448538
- PMCID: PMC2447050
- DOI: 10.1128/JVI.00554-08
Comparison of G-to-A mutation frequencies induced by APOBEC3 proteins in H9 cells and peripheral blood mononuclear cells in the context of impaired processivities of drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants
Abstract
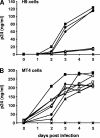
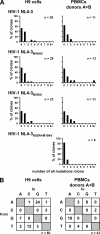
APOBEC3 proteins can inhibit human immunodeficiency virus type 1 (HIV-1) replication by inducing G-to-A mutations in newly synthesized viral DNA. However, HIV-1 is able to overcome the antiretroviral activity of some of those enzymes by the viral protein Vif. We investigated the impact of different processivities of HIV-1 reverse transcriptases (RT) on the frequencies of G-to-A mutations introduced by APOBEC3 proteins. Wild-type RT or the M184V, M184I, and K65R+M184V RT variants, which are increasingly impaired in their processivities, were used in the context of a vif-deficient molecular HIV-1 clone to infect H9 cells and peripheral blood mononuclear cells (PBMCs). After two rounds of infection, a part of the HIV-1 env gene was amplified, cloned, and sequenced. The M184V mutation led to G-to-A mutation frequencies that were similar to those of the wild-type RT in H9 cells and PBMCs. The frequencies of G-to-A mutations were increased after infection with the M184I virus variant. This effect was augmented when using the K65R+M184V virus variant (P < 0.001). Overall, the G-to-A mutation frequencies were lower in PBMCs than in H9 cells. Remarkably, 38% +/- 18% (mean +/- standard deviation) of the env clones derived from PBMCs did not harbor any G-to-A mutation. This was rarely observed in H9 cells (3% +/- 3%). Our data imply that the frequency of G-to-A mutations induced by APOBEC3 proteins can be influenced by the processivities of HIV-1 RT variants. The high number of nonmutated clones derived from PBMCs leads to several hypotheses, including that additional antiretroviral mechanisms of APOBEC3 proteins other than their deamination activity might be involved in the inhibition of vif-deficient viruses.
Figures

Similar articles
-
E138K and M184I mutations in HIV-1 reverse transcriptase coemerge as a result of APOBEC3 editing in the absence of drug exposure.AIDS. 2012 Aug 24;26(13):1619-24. doi: 10.1097/QAD.0b013e3283560703. AIDS. 2012. PMID: 22695298
-
In-depth analysis of G-to-A hypermutation rate in HIV-1 env DNA induced by endogenous APOBEC3 proteins using massively parallel sequencing.J Virol Methods. 2011 Feb;171(2):329-38. doi: 10.1016/j.jviromet.2010.11.016. Epub 2010 Nov 24. J Virol Methods. 2011. PMID: 21111003
-
Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity.Antimicrob Agents Chemother. 2002 Nov;46(11):3437-46. doi: 10.1128/AAC.46.11.3437-3446.2002. Antimicrob Agents Chemother. 2002. PMID: 12384348 Free PMC article.
-
Interaction between HIV-1 and APOBEC3 sub-family of proteins.Curr HIV Res. 2006 Oct;4(4):401-9. doi: 10.2174/157016206778560063. Curr HIV Res. 2006. PMID: 17073615 Review.
-
Powerful mutators lurking in the genome.Philos Trans R Soc Lond B Biol Sci. 2009 Mar 12;364(1517):705-15. doi: 10.1098/rstb.2008.0272. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19042181 Free PMC article. Review.
Cited by
-
Vif substitution enables persistent infection of pig-tailed macaques by human immunodeficiency virus type 1.J Virol. 2011 Apr;85(8):3767-79. doi: 10.1128/JVI.02438-10. Epub 2011 Feb 2. J Virol. 2011. PMID: 21289128 Free PMC article.
-
Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets.J Virol. 2009 Sep;83(18):9474-85. doi: 10.1128/JVI.01089-09. Epub 2009 Jul 8. J Virol. 2009. PMID: 19587057 Free PMC article.
-
Exposure to apoptotic activated CD4+ T cells induces maturation and APOBEC3G-mediated inhibition of HIV-1 infection in dendritic cells.PLoS One. 2011;6(6):e21171. doi: 10.1371/journal.pone.0021171. Epub 2011 Jun 16. PLoS One. 2011. PMID: 21698207 Free PMC article.
-
BST2 Suppresses LINE-1 Retrotransposition by Reducing the Promoter Activity of LINE-1 5' UTR.J Virol. 2022 Jan 26;96(2):e0161021. doi: 10.1128/JVI.01610-21. Epub 2021 Nov 3. J Virol. 2022. PMID: 34730388 Free PMC article.
-
APOBEC3G inhibits elongation of HIV-1 reverse transcripts.PLoS Pathog. 2008 Dec;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. Epub 2008 Dec 5. PLoS Pathog. 2008. PMID: 19057663 Free PMC article.
References
-
- Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 27934083-34086. - PubMed
-
- Allers, K., S. A. Knoepfel, P. Rauch, H. Walter, M. Opravil, M. Fischer, H. F. Gunthard, and K. J. Metzner. 2007. Persistence of lamivudine-sensitive HIV-1 quasispecies in the presence of lamivudine in vitro and in vivo. J. Acquir. Immune Defic. Syndr. 44377-385. - PubMed
-
- Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 154040-4049. - PMC - PubMed
-
- Beale, R. C., S. K. Petersen-Mahrt, I. N. Watt, R. S. Harris, C. Rada, and M. S. Neuberger. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337585-596. - PubMed
-
- Berkhout, B., and A. de Ronde. 2004. APOBEC3G versus reverse transcriptase in the generation of HIV-1 drug-resistance mutations. AIDS 181861-1863. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

