Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction
- PMID: 18446221
- PMCID: PMC2323571
- DOI: 10.1371/journal.pone.0002032
Processing of genome 5' termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction
Abstract
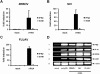
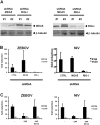
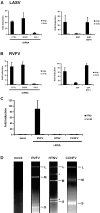
Innate immunity is critically dependent on the rapid production of interferon in response to intruding viruses. The intracellular pathogen recognition receptors RIG-I and MDA5 are essential for interferon induction by viral RNAs containing 5' triphosphates or double-stranded structures, respectively. Viruses with a negative-stranded RNA genome are an important group of pathogens causing emerging and re-emerging diseases. We investigated the ability of genomic RNAs from substantial representatives of this virus group to induce interferon via RIG-I or MDA5. RNAs isolated from particles of Ebola virus, Nipah virus, Lassa virus, and Rift Valley fever virus strongly activated the interferon-beta promoter. Knockdown experiments demonstrated that interferon induction depended on RIG-I, but not MDA5, and phosphatase treatment revealed a requirement for the RNA 5' triphosphate group. In contrast, genomic RNAs of Hantaan virus, Crimean-Congo hemorrhagic fever virus and Borna disease virus did not trigger interferon induction. Sensitivity of these RNAs to a 5' monophosphate-specific exonuclease indicates that the RIG-I-activating 5' triphosphate group was removed post-transcriptionally by a viral function. Consequently, RIG-I is unable to bind the RNAs of Hantaan virus, Crimean-Congo hemorrhagic fever virus and Borna disease virus. These results establish RIG-I as a major intracellular recognition receptor for the genome of most negative-strand RNA viruses and define the cleavage of triphosphates at the RNA 5' end as a strategy of viruses to evade the innate immune response.
Conflict of interest statement
Figures

Similar articles
-
Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit.Antiviral Res. 2013 Dec;100(3):615-35. doi: 10.1016/j.antiviral.2013.10.002. Epub 2013 Oct 12. Antiviral Res. 2013. PMID: 24129118 Free PMC article. Review.
-
RIG-I detects viral genomic RNA during negative-strand RNA virus infection.Cell. 2010 Feb 5;140(3):397-408. doi: 10.1016/j.cell.2010.01.020. Cell. 2010. PMID: 20144762
-
RIG-I Mediates an Antiviral Response to Crimean-Congo Hemorrhagic Fever Virus.J Virol. 2015 Oct;89(20):10219-29. doi: 10.1128/JVI.01643-15. Epub 2015 Jul 29. J Virol. 2015. PMID: 26223644 Free PMC article.
-
RIG-I-like receptors and negative-strand RNA viruses: RLRly bird catches some worms.Cytokine Growth Factor Rev. 2014 Oct;25(5):621-8. doi: 10.1016/j.cytogfr.2014.05.004. Epub 2014 May 20. Cytokine Growth Factor Rev. 2014. PMID: 24894317 Free PMC article. Review.
-
Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses.Nature. 2006 May 4;441(7089):101-5. doi: 10.1038/nature04734. Epub 2006 Apr 9. Nature. 2006. PMID: 16625202
Cited by
-
Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response.PLoS One. 2012;7(10):e47790. doi: 10.1371/journal.pone.0047790. Epub 2012 Oct 19. PLoS One. 2012. PMID: 23094089 Free PMC article.
-
The chase for the RIG-I ligand--recent advances.Mol Ther. 2010 Jul;18(7):1254-62. doi: 10.1038/mt.2010.90. Epub 2010 May 11. Mol Ther. 2010. PMID: 20461060 Free PMC article. Review.
-
Genomic RNAs of Borna disease virus are elongated on internal template motifs after realignment of the 3' termini.Proc Natl Acad Sci U S A. 2011 Apr 26;108(17):7206-11. doi: 10.1073/pnas.1016759108. Epub 2011 Apr 11. Proc Natl Acad Sci U S A. 2011. PMID: 21482759 Free PMC article.
-
Cell entry of lymphocytic choriomeningitis virus is restricted in myotubes.Virology. 2014 Jun;458-459:22-32. doi: 10.1016/j.virol.2014.04.013. Epub 2014 May 5. Virology. 2014. PMID: 24928036 Free PMC article.
-
RNA helicase signaling is critical for type i interferon production and protection against Rift Valley fever virus during mucosal challenge.J Virol. 2013 May;87(9):4846-60. doi: 10.1128/JVI.01997-12. Epub 2013 Feb 13. J Virol. 2013. PMID: 23408632 Free PMC article.
References
-
- Bowie AG, Fitzgerald KA. RIG-I: tri-ing to discriminate between self and non-self RNA. Trends Immunol 2007 - PubMed
-
- Pichlmair A, Reis ESC. Innate Recognition of Viruses. Immunity. 2007;27:370–383. - PubMed
-
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. - PubMed
-
- Garcia-Sastre A, Biron CA. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

