Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians
- PMID: 18439545
- PMCID: PMC2427302
- DOI: 10.1016/j.ajhg.2008.03.012
Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians
Abstract
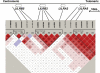
Human leukocyte antigen (HLA) plays a critical role in innate and adaptive immunity and is a well-known example of genes under natural selection. However, the genetic aspect of receptors recognizing HLA molecules has not yet been fully elucidated. Leukocyte immunoglobulin (Ig)-like receptors (LILRs) are a family of HLA class I-recognizing receptors comprising activating and inhibitory forms. We previously reported that the allele frequency of the 6.7 kb LILRA3 deletion is extremely high (71%) in the Japanese population, and we identified premature termination codon (PTC)-containing alleles. In this study, we observed a wide distribution of the high deletion frequency in Northeast Asians (84% in Korean Chinese, 79% in Man Chinese, 56% in Mongolian, and 76% in Buryat populations). Genotyping of the four HapMap populations revealed that LILRA3 alleles were in strong linkage disequilibrium with LILRB2 alleles in Northeast Asians. In addition, PTC-containing LILRA3 alleles were detected in Northeast Asians but not in non-Northeast Asians. Furthermore, flow-cytometric analysis revealed that the LILRB2 allele frequent in Northeast Asians was significantly associated with low levels of expression. F(ST) and extended-haplotype-homozygosity analysis for the HapMap populations provided evidence of positive selection acting on the LILRA3 and LILRB2 loci. Taken together, our results suggest that both the nonfunctional LILRA3 alleles and the low-expressing LILRB2 alleles identified in our study have increased in Northeast Asians because of natural selection. Our findings, therefore, lead us to speculate that not only HLA class I ligands but also their receptors might be sensitive to the local environment.
Figures


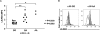
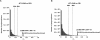

Similar articles
-
LILRB2 interaction with HLA class I correlates with control of HIV-1 infection.PLoS Genet. 2014 Mar 6;10(3):e1004196. doi: 10.1371/journal.pgen.1004196. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603468 Free PMC article.
-
Genetic link between Asians and native Americans: evidence from HLA genes and haplotypes.Hum Immunol. 2001 Sep;62(9):1001-8. doi: 10.1016/s0198-8859(01)00301-9. Hum Immunol. 2001. PMID: 11543902
-
HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding.J Immunol. 2011 Mar 1;186(5):2990-7. doi: 10.4049/jimmunol.1003078. Epub 2011 Jan 26. J Immunol. 2011. PMID: 21270408
-
Genetic susceptibility factors of Type 1 diabetes in Asians.Diabetes Metab Res Rev. 2001 Jan-Feb;17(1):2-11. doi: 10.1002/1520-7560(2000)9999:9999<::aid-dmrr164>3.0.co;2-m. Diabetes Metab Res Rev. 2001. PMID: 11241886 Review.
-
Evolution and molecular interactions of major histocompatibility complex (MHC)-G, -E and -F genes.Cell Mol Life Sci. 2022 Aug 4;79(8):464. doi: 10.1007/s00018-022-04491-z. Cell Mol Life Sci. 2022. PMID: 35925520 Free PMC article. Review.
Cited by
-
Fibrinogen induces inflammatory responses via the immune activating receptor LILRA2.Front Immunol. 2024 Sep 23;15:1435236. doi: 10.3389/fimmu.2024.1435236. eCollection 2024. Front Immunol. 2024. PMID: 39376567 Free PMC article.
-
Genome-Wide Association Analysis Identifies LILRB2 Gene for Pathological Myopia.Adv Sci (Weinh). 2024 Oct;11(40):e2308968. doi: 10.1002/advs.202308968. Epub 2024 Aug 29. Adv Sci (Weinh). 2024. PMID: 39207058 Free PMC article.
-
Identification of the hybrid gene LILRB5-3 by long-read sequencing and implication of its novel signaling function.Front Immunol. 2024 May 14;15:1398935. doi: 10.3389/fimmu.2024.1398935. eCollection 2024. Front Immunol. 2024. PMID: 38807600 Free PMC article.
-
Population-enriched innate immune variants may identify candidate gene targets at the intersection of cancer and cardio-metabolic disease.Front Endocrinol (Lausanne). 2024 Mar 21;14:1286979. doi: 10.3389/fendo.2023.1286979. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38577257 Free PMC article. Review.
-
Immune receptors and aging brain.Biosci Rep. 2024 Feb 29;44(2):BSR20222267. doi: 10.1042/BSR20222267. Biosci Rep. 2024. PMID: 38299364 Free PMC article. Review.
References
-
- Meyer D., Thomson G. How selection shapes variation of the human major histocompatibility complex: A review. Ann. Hum. Genet. 2001;65:1–26. - PubMed
-
- Brown D., Trowsdale J., Allen R. The LILR family: Modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–225. - PubMed
-
- Colonna M., Nakajima H., Navarro F., Lopez-Botet M. A novel family of Ig-like receptors for HLA class I molecules that modulate function of lymphoid and myeloid cells. J. Leukoc. Biol. 1999;66:375–381. - PubMed
-
- Fanger N.A., Borges L., Cosman D. The leukocyte immunoglobulin-like receptors (LIRs): A new family of immune regulators. J. Leukoc. Biol. 1999;66:231–236. - PubMed
-
- Heinzmann A., Blattmann S., Forster J., Kuehr J., Deichmann K.A. Common polymorphisms and alternative splicing in the ILT3 gene are not associated with atopy. Eur. J. Immunogenet. 2000;27:121–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

