The many faces of ubiquitinated histone H2A: insights from the DUBs
- PMID: 18430235
- PMCID: PMC2373781
- DOI: 10.1186/1747-1028-3-8
The many faces of ubiquitinated histone H2A: insights from the DUBs
Abstract
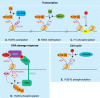
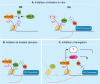
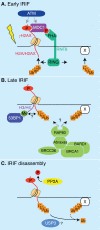
Monoubiquitination of H2A is a major histone modification in mammalian cells. Understanding how monoubiquitinated H2A (uH2A) regulates DNA-based processes in the context of chromatin is a challenging question. Work in the past years linked uH2A to transcriptional repression by the Polycomb group proteins of developmental regulators. Recently, a number of mammalian deubiquitinating enzymes (DUBs) that catalyze the removal of ubiquitin from H2A have been discovered. These studies provide convincing evidence that H2A deubiquitination is connected with gene activation. In addition, uH2A regulatory enzymes have crucial roles in the cellular response to DNA damage and in cell cycle progression. In this review we will discuss new insights into uH2A biology, with emphasis on the H2A DUBs.
Figures
Similar articles
-
Caspase-dependent deubiquitination of monoubiquitinated nucleosomal histone H2A induced by diverse apoptogenic stimuli.Cell Death Differ. 2001 Dec;8(12):1182-96. doi: 10.1038/sj.cdd.4400924. Cell Death Differ. 2001. PMID: 11753566
-
Human USP3 is a chromatin modifier required for S phase progression and genome stability.Curr Biol. 2007 Nov 20;17(22):1972-7. doi: 10.1016/j.cub.2007.10.034. Epub 2007 Nov 1. Curr Biol. 2007. PMID: 17980597
-
H2A-DUBbing the mammalian epigenome: expanding frontiers for histone H2A deubiquitinating enzymes in cell biology and physiology.Int J Biochem Cell Biol. 2014 May;50:161-74. doi: 10.1016/j.biocel.2014.03.004. Epub 2014 Mar 16. Int J Biochem Cell Biol. 2014. PMID: 24647359 Review.
-
Fine-tuning the ubiquitin code at DNA double-strand breaks: deubiquitinating enzymes at work.Front Genet. 2015 Sep 8;6:282. doi: 10.3389/fgene.2015.00282. eCollection 2015. Front Genet. 2015. PMID: 26442100 Free PMC article. Review.
-
Histone H2A monoubiquitination and Polycomb repression: the missing pieces of the puzzle.Fly (Austin). 2012 Jul-Sep;6(3):162-8. doi: 10.4161/fly.20986. Epub 2012 Jul 1. Fly (Austin). 2012. PMID: 22836728
Cited by
-
Ubiquitination dynamics in the early-branching eukaryote Giardia intestinalis.Microbiologyopen. 2013 Jun;2(3):525-39. doi: 10.1002/mbo3.88. Epub 2013 Apr 23. Microbiologyopen. 2013. PMID: 23613346 Free PMC article.
-
The ubiquitin proteasome system in neuropathology.Acta Neuropathol. 2009 Sep;118(3):329-47. doi: 10.1007/s00401-009-0560-x. Epub 2009 Jul 14. Acta Neuropathol. 2009. PMID: 19597829 Free PMC article. Review.
-
A conserved function for the H2A.Z C terminus.J Biol Chem. 2012 Jun 1;287(23):19148-57. doi: 10.1074/jbc.M111.317990. Epub 2012 Apr 9. J Biol Chem. 2012. PMID: 22493515 Free PMC article.
-
Ubiquitin and the DNA damage response: a new handle on histones.Cell Cycle. 2012 Sep 1;11(17):3153. doi: 10.4161/cc.21718. Epub 2012 Aug 16. Cell Cycle. 2012. PMID: 22895171 Free PMC article. No abstract available.
-
Atypical Ubiquitination and Parkinson's Disease.Int J Mol Sci. 2022 Mar 28;23(7):3705. doi: 10.3390/ijms23073705. Int J Mol Sci. 2022. PMID: 35409068 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

