The Catalogue of Somatic Mutations in Cancer (COSMIC)
- PMID: 18428421
- PMCID: PMC2705836
- DOI: 10.1002/0471142905.hg1011s57
The Catalogue of Somatic Mutations in Cancer (COSMIC)
Abstract
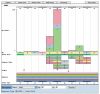
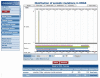
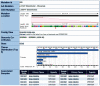
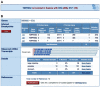
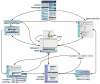
COSMIC is currently the most comprehensive global resource for information on somatic mutations in human cancer, combining curation of the scientific literature with tumor resequencing data from the Cancer Genome Project at the Sanger Institute, U.K. Almost 4800 genes and 250000 tumors have been examined, resulting in over 50000 mutations available for investigation. This information can be accessed in a number of ways, the most convenient being the Web-based system which allows detailed data mining, presenting the results in easily interpretable formats. This unit describes the graphical system in detail, elaborating an example walkthrough and the many ways that the resulting information can be thoroughly investigated by combining data, respecializing the query, or viewing the results in different ways. Alternate protocols overview the available precompiled data files available for download.
Copyright 2008 by John Wiley & Sons, Inc.
Figures









Similar articles
-
COSMIC: High-Resolution Cancer Genetics Using the Catalogue of Somatic Mutations in Cancer.Curr Protoc Hum Genet. 2016 Oct 11;91:10.11.1-10.11.37. doi: 10.1002/cphg.21. Curr Protoc Hum Genet. 2016. PMID: 27727438
-
COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer.Nucleic Acids Res. 2010 Jan;38(Database issue):D652-7. doi: 10.1093/nar/gkp995. Epub 2009 Nov 11. Nucleic Acids Res. 2010. PMID: 19906727 Free PMC article.
-
COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer.Nucleic Acids Res. 2011 Jan;39(Database issue):D945-50. doi: 10.1093/nar/gkq929. Epub 2010 Oct 15. Nucleic Acids Res. 2011. PMID: 20952405 Free PMC article.
-
Combinatorial patterns of somatic gene mutations in cancer.FASEB J. 2008 Aug;22(8):2605-22. doi: 10.1096/fj.08-108985. Epub 2008 Apr 23. FASEB J. 2008. PMID: 18434431 Review.
-
The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers.Nat Rev Cancer. 2018 Nov;18(11):696-705. doi: 10.1038/s41568-018-0060-1. Nat Rev Cancer. 2018. PMID: 30293088 Free PMC article. Review.
Cited by
-
Mutated genes and driver pathways involved in myelodysplastic syndromes—a transcriptome sequencing based approach.Mol Biosyst. 2015 Aug;11(8):2158-66. doi: 10.1039/c4mb00663a. Mol Biosyst. 2015. PMID: 26010722 Free PMC article.
-
JAFFA: High sensitivity transcriptome-focused fusion gene detection.Genome Med. 2015 May 11;7(1):43. doi: 10.1186/s13073-015-0167-x. eCollection 2015. Genome Med. 2015. PMID: 26019724 Free PMC article.
-
N-glycosylation controls the function of junctional adhesion molecule-A.Mol Biol Cell. 2015 Sep 15;26(18):3205-14. doi: 10.1091/mbc.E14-12-1604. Epub 2015 Jul 29. Mol Biol Cell. 2015. PMID: 26224316 Free PMC article.
-
Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia.Nat Commun. 2016 Jun 6;7:11790. doi: 10.1038/ncomms11790. Nat Commun. 2016. PMID: 27265895 Free PMC article.
-
The dJ/dS Ratio Test Reveals Hundreds of Novel Putative Cancer Drivers.Mol Biol Evol. 2015 Aug;32(8):2181-5. doi: 10.1093/molbev/msv083. Epub 2015 Apr 14. Mol Biol Evol. 2015. PMID: 25873590 Free PMC article.
References
Literature Cited
-
- Cooper DN, Stenson PD, Chuzhanove NA. The human gene mutation database (HGMD) and its exploitation in the study of human mutational mechanisms. Curr. Protoc. Bioinform. 2005;12:1.13.1–1.13.20. - PubMed
-
- denDunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum. Mutat. 2000;15:7–12. - PubMed
-
- Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, Hunter C, Smith R, Stephens P, Goldstraw P, Nicholson A, Chan TL, Velculescu VE, Yuen ST, Leung SY, Stratton MR, Futreal PA. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol. Ther. 2006;5:928–932. - PMC - PubMed
Internet Resources
-
- COSMIC home page. http://www.sanger.ac.uk/cosmic.
-
- Ensembl home page. http://www.ensembl.org.
-
- Cancer Gene Census. http://www.sanger.ac.uk/genetics/cgp/census.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

