Using genetic markers to orient the edges in quantitative trait networks: the NEO software
- PMID: 18412962
- PMCID: PMC2387136
- DOI: 10.1186/1752-0509-2-34
Using genetic markers to orient the edges in quantitative trait networks: the NEO software
Abstract
Background: Systems genetic studies have been used to identify genetic loci that affect transcript abundances and clinical traits such as body weight. The pairwise correlations between gene expression traits and/or clinical traits can be used to define undirected trait networks. Several authors have argued that genetic markers (e.g expression quantitative trait loci, eQTLs) can serve as causal anchors for orienting the edges of a trait network. The availability of hundreds of thousands of genetic markers poses new challenges: how to relate (anchor) traits to multiple genetic markers, how to score the genetic evidence in favor of an edge orientation, and how to weigh the information from multiple markers.
Results: We develop and implement Network Edge Orienting (NEO) methods and software that address the challenges of inferring unconfounded and directed gene networks from microarray-derived gene expression data by integrating mRNA levels with genetic marker data and Structural Equation Model (SEM) comparisons. The NEO software implements several manual and automatic methods for incorporating genetic information to anchor traits. The networks are oriented by considering each edge separately, thus reducing error propagation. To summarize the genetic evidence in favor of a given edge orientation, we propose Local SEM-based Edge Orienting (LEO) scores that compare the fit of several competing causal graphs. SEM fitting indices allow the user to assess local and overall model fit. The NEO software allows the user to carry out a robustness analysis with regard to genetic marker selection. We demonstrate the utility of NEO by recovering known causal relationships in the sterol homeostasis pathway using liver gene expression data from an F2 mouse cross. Further, we use NEO to study the relationship between a disease gene and a biologically important gene co-expression module in liver tissue.
Conclusion: The NEO software can be used to orient the edges of gene co-expression networks or quantitative trait networks if the edges can be anchored to genetic marker data. R software tutorials, data, and supplementary material can be downloaded from: http://www.genetics.ucla.edu/labs/horvath/aten/NEO.
Figures
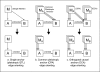
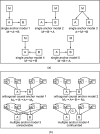

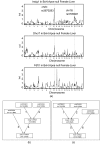
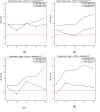
Similar articles
-
FunMap: functional mapping of complex traits.Bioinformatics. 2004 Jul 22;20(11):1808-11. doi: 10.1093/bioinformatics/bth156. Epub 2004 Feb 26. Bioinformatics. 2004. PMID: 14988108
-
R/qtl: QTL mapping in experimental crosses.Bioinformatics. 2003 May 1;19(7):889-90. doi: 10.1093/bioinformatics/btg112. Bioinformatics. 2003. PMID: 12724300
-
Eigengene networks for studying the relationships between co-expression modules.BMC Syst Biol. 2007 Nov 21;1:54. doi: 10.1186/1752-0509-1-54. BMC Syst Biol. 2007. PMID: 18031580 Free PMC article.
-
eQTL analysis in mice and rats.Methods Mol Biol. 2009;573:285-309. doi: 10.1007/978-1-60761-247-6_16. Methods Mol Biol. 2009. PMID: 19763934 Review.
-
Moving toward a system genetics view of disease.Mamm Genome. 2007 Jul;18(6-7):389-401. doi: 10.1007/s00335-007-9040-6. Epub 2007 Jul 26. Mamm Genome. 2007. PMID: 17653589 Free PMC article. Review.
Cited by
-
Systems genetic analysis of osteoblast-lineage cells.PLoS Genet. 2012;8(12):e1003150. doi: 10.1371/journal.pgen.1003150. Epub 2012 Dec 27. PLoS Genet. 2012. PMID: 23300464 Free PMC article.
-
Elucidating molecular networks that either affect or respond to plasma cortisol concentration in target tissues of liver and muscle.Genetics. 2012 Nov;192(3):1109-22. doi: 10.1534/genetics.112.143081. Epub 2012 Aug 17. Genetics. 2012. PMID: 22904034 Free PMC article.
-
The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders.Sci Transl Med. 2018 Dec 19;10(472):eaat8178. doi: 10.1126/scitranslmed.aat8178. Epub 2018 Dec 13. Sci Transl Med. 2018. PMID: 30545964 Free PMC article.
-
Learning gene networks under SNP perturbations using eQTL datasets.PLoS Comput Biol. 2014 Feb 27;10(2):e1003420. doi: 10.1371/journal.pcbi.1003420. eCollection 2014 Feb. PLoS Comput Biol. 2014. PMID: 24586125 Free PMC article.
-
Bicc1 is a genetic determinant of osteoblastogenesis and bone mineral density.J Clin Invest. 2014 Jun;124(6):2736-49. doi: 10.1172/JCI73072. Epub 2014 May 1. J Clin Invest. 2014. PMID: 24789909 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

