Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos
- PMID: 18410726
- PMCID: PMC2901920
- DOI: 10.1016/j.devcel.2008.01.018
Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos
Abstract
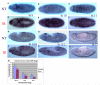
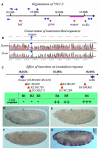
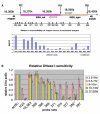
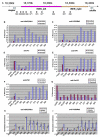
Drosophila embryos are highly sensitive to gamma-ray-induced apoptosis at early but not later, more differentiated stages during development. Two proapoptotic genes, reaper and hid, are upregulated rapidly following irradiation. However, in post-stage-12 embryos, in which most cells have begun differentiation, neither proapoptotic gene can be induced by high doses of irradiation. Our study indicates that the sensitive-to-resistant transition is due to epigenetic blocking of the irradiation-responsive enhancer region (IRER), which is located upstream of reaper but is also required for the induction of hid in response to irradiation. This IRER, but not the transcribed regions of reaper/hid, becomes enriched for trimethylated H3K27/H3K9 and forms a heterochromatin-like structure during the sensitive-to-resistant transition. The functions of histone-modifying enzymes Hdac1(rpd3) and Su(var)3-9 and PcG proteins Su(z)12 and Polycomb are required for this process. Thus, direct epigenetic regulation of two proapoptotic genes controls cellular sensitivity to cytotoxic stimuli.
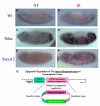
Figures


Similar articles
-
In vivo analysis of Drosophila SU(Z)12 function.Mol Genet Genomics. 2008 Feb;279(2):159-70. doi: 10.1007/s00438-007-0304-3. Epub 2007 Nov 22. Mol Genet Genomics. 2008. PMID: 18034266
-
A barrier-only boundary element delimits the formation of facultative heterochromatin in Drosophila melanogaster and vertebrates.Mol Cell Biol. 2011 Jul;31(13):2729-41. doi: 10.1128/MCB.05165-11. Epub 2011 Apr 25. Mol Cell Biol. 2011. PMID: 21518956 Free PMC article.
-
SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex.Curr Biol. 2004 Oct 26;14(20):1812-21. doi: 10.1016/j.cub.2004.09.060. Curr Biol. 2004. PMID: 15498488
-
Histone modification and the control of heterochromatic gene silencing in Drosophila.Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
-
Recruiting polycomb to chromatin.Int J Biochem Cell Biol. 2015 Oct;67:177-87. doi: 10.1016/j.biocel.2015.05.006. Epub 2015 May 14. Int J Biochem Cell Biol. 2015. PMID: 25982201 Free PMC article. Review.
Cited by
-
Functional association between eyegone and HP1a mediates wingless transcriptional repression during development.Mol Cell Biol. 2012 Jul;32(13):2407-15. doi: 10.1128/MCB.06311-11. Epub 2012 Apr 30. Mol Cell Biol. 2012. PMID: 22547675 Free PMC article.
-
Induction of reaper ortholog mx in mosquito midgut cells following baculovirus infection.Cell Death Differ. 2011 Aug;18(8):1337-45. doi: 10.1038/cdd.2011.8. Epub 2011 Feb 18. Cell Death Differ. 2011. PMID: 21331076 Free PMC article.
-
Induction of rapid and selective cell necrosis in Drosophila using Bacillus thuringiensis Cry toxin and its silkworm receptor.BMC Biol. 2015 Jul 8;13:48. doi: 10.1186/s12915-015-0160-2. BMC Biol. 2015. PMID: 26152191 Free PMC article.
-
In vivo monitoring of stem cells in Drosophila pupae using the radiative transfer equation-based fluorescence molecular tomography.Mol Imaging Biol. 2011 Oct;13(5):868-73. doi: 10.1007/s11307-010-0434-6. Mol Imaging Biol. 2011. PMID: 20922571
-
Genetic control of programmed cell death during animal development.Annu Rev Genet. 2009;43:493-523. doi: 10.1146/annurev.genet.42.110807.091533. Annu Rev Genet. 2009. PMID: 19886811 Free PMC article. Review.
References
-
- Ashburner M. A Laboratory Handbook. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. Drosophila.
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer– a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. - PubMed
-
- Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

