Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system
- PMID: 18407958
- PMCID: PMC2493377
- DOI: 10.1074/mcp.M800079-MCP200
Monitoring protein-protein interactions between the mammalian integral membrane transporters and PDZ-interacting partners using a modified split-ubiquitin membrane yeast two-hybrid system
Abstract
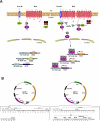
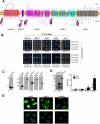
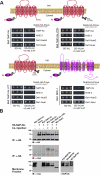
PDZ-binding motifs are found in the C-terminal tails of numerous integral membrane proteins where they mediate specific protein-protein interactions by binding to PDZ-containing proteins. Conventional yeast two-hybrid screens have been used to probe protein-protein interactions of these soluble C termini. However, to date no in vivo technology has been available to study interactions between the full-length integral membrane proteins and their cognate PDZ-interacting partners. We previously developed a split-ubiquitin membrane yeast two-hybrid (MYTH) system to test interactions between such integral membrane proteins by using a transcriptional output based on cleavage of a transcription factor from the C terminus of membrane-inserted baits. Here we modified MYTH to permit detection of C-terminal PDZ domain interactions by redirecting the transcription factor moiety from the C to the N terminus of a given integral membrane protein thus liberating their native C termini. We successfully applied this "MYTH 2.0" system to five different mammalian full-length renal transporters and identified novel PDZ domain-containing partners of the phosphate (NaPi-IIa) and sulfate (NaS1) transporters that would have otherwise not been detectable. Furthermore this assay was applied to locate the PDZ-binding domain on the NaS1 protein. We showed that the PDZ-binding domain for PDZK1 on NaS1 is upstream of its C terminus, whereas the two interacting proteins, NHERF-1 and NHERF-2, bind at a location closer to the N terminus of NaS1. Moreover NHERF-1 and NHERF-2 increased functional sulfate uptake in Xenopus oocytes when co-expressed with NaS1. Finally we used MYTH 2.0 to demonstrate that the NaPi-IIa transporter homodimerizes via protein-protein interactions within the lipid bilayer. In summary, our study establishes the MYTH 2.0 system as a novel tool for interactive proteomics studies of membrane protein complexes.
Figures



Similar articles
-
PDZK1: I. a major scaffolder in brush borders of proximal tubular cells.Kidney Int. 2003 Nov;64(5):1733-45. doi: 10.1046/j.1523-1755.2003.00266.x. Kidney Int. 2003. PMID: 14531806
-
Split-ubiquitin based membrane yeast two-hybrid (MYTH) system: a powerful tool for identifying protein-protein interactions.J Vis Exp. 2010 Feb 1;(36):1698. doi: 10.3791/1698. J Vis Exp. 2010. PMID: 20125081 Free PMC article.
-
PDZK1: II. an anchoring site for the PKA-binding protein D-AKAP2 in renal proximal tubular cells.Kidney Int. 2003 Nov;64(5):1746-54. doi: 10.1046/j.1523-1755.2003.00267.x. Kidney Int. 2003. PMID: 14531807
-
The split-ubiquitin membrane-based yeast two-hybrid system.Methods Mol Biol. 2004;261:297-312. doi: 10.1385/1-59259-762-9:297. Methods Mol Biol. 2004. PMID: 15064465 Review.
-
Using yeast as a model to study membrane proteins.Curr Opin Nephrol Hypertens. 2011 Jul;20(4):425-32. doi: 10.1097/MNH.0b013e3283478611. Curr Opin Nephrol Hypertens. 2011. PMID: 21587075 Review.
Cited by
-
Systematic protein-protein interaction mapping for clinically relevant human GPCRs.Mol Syst Biol. 2017 Mar 15;13(3):918. doi: 10.15252/msb.20167430. Mol Syst Biol. 2017. PMID: 28298427 Free PMC article.
-
Molecular Studies of the Protein Complexes Involving Cis-Prenyltransferase in Guayule (Parthenium argentatum), an Alternative Rubber-Producing Plant.Front Plant Sci. 2019 Feb 25;10:165. doi: 10.3389/fpls.2019.00165. eCollection 2019. Front Plant Sci. 2019. PMID: 30858856 Free PMC article.
-
ABC transporters in Saccharomyces cerevisiae and their interactors: new technology advances the biology of the ABCC (MRP) subfamily.Microbiol Mol Biol Rev. 2009 Dec;73(4):577-93. doi: 10.1128/MMBR.00020-09. Microbiol Mol Biol Rev. 2009. PMID: 19946134 Free PMC article. Review.
-
Protein-protein interactions: switch from classical methods to proteomics and bioinformatics-based approaches.Cell Mol Life Sci. 2014 Jan;71(2):205-28. doi: 10.1007/s00018-013-1333-1. Epub 2013 Apr 12. Cell Mol Life Sci. 2014. PMID: 23579629 Free PMC article. Review.
-
Structural basis for NHERF1 PDZ domain binding.Biochemistry. 2012 Apr 10;51(14):3110-20. doi: 10.1021/bi201213w. Epub 2012 Mar 27. Biochemistry. 2012. PMID: 22429102 Free PMC article.
References
-
- Hung A. Y., and Sheng, M. ( 2002) PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277, 5699–5702 - PubMed
-
- Hillier, B. J., Christopherson, K. S., Prehoda, K. E., Bredt, D. S., and Lim, W. A. ( 1999) Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science 284, 812–815 - PubMed
-
- Bezprozvanny, I., and Maximov, A. ( 2001) Classification of PDZ domains. FEBS Lett. 509, 457–462 - PubMed
-
- Hernando, N., Wagner, C. A., Gisler, S. M., Biber, J., and Murer, H. ( 2004) PDZ proteins and proximal ion transport. Curr. Opin. Nephrol. Hypertens. 13, 569–574 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

