Altered histone monoubiquitylation mediated by mutant huntingtin induces transcriptional dysregulation
- PMID: 18400894
- PMCID: PMC3844822
- DOI: 10.1523/JNEUROSCI.5667-07.2008
Altered histone monoubiquitylation mediated by mutant huntingtin induces transcriptional dysregulation
Abstract
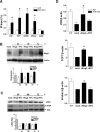
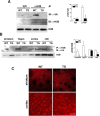
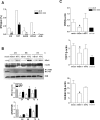
Although transcriptional dysregulation is a critical pathogenic mechanism in Huntington's disease (HD), it is still not known how mutant huntingtin causes it. Here we show that alteration of histone monoubiquitylation is a key mechanism. Disrupted interaction of huntingtin with Bmi-1, a component of the hPRC1L E3 ubiquitin ligase complex, increases monoubiquityl histone H2A (uH2A) levels in a cell culture model of HD. Genes with expression that is repressed in transgenic R6/2 mouse brain have increased uH2A and decreased uH2B at their promoters, whereas actively transcribed genes show the opposite pattern. Reduction in uH2A reverses transcriptional repression and inhibits methylation of histone H3 at lysine 9 in cell culture. In contrast, reduction in uH2B induces transcriptional repression and inhibits methylation of histone H3 at lysine 4. This is the first report to demonstrate hPRC1L as a huntingtin-interacting histone modifying complex and a crucial role for histone monoubiquitylation in mammalian brain gene expression, which broadens our understanding of histone code. These findings also provide a rationale for targeting histone monoubiquitylation for therapy in HD.
Figures





Similar articles
-
Genome-wide increase in histone H2A ubiquitylation in a mouse model of Huntington's disease.J Huntingtons Dis. 2013;2(3):263-77. doi: 10.3233/JHD-130066. J Huntingtons Dis. 2013. PMID: 25062675
-
Modulation of nucleosome dynamics in Huntington's disease.Hum Mol Genet. 2007 May 15;16(10):1164-75. doi: 10.1093/hmg/ddm064. Epub 2007 Apr 2. Hum Mol Genet. 2007. PMID: 17403718
-
Genome-wide uH2A localization analysis highlights Bmi1-dependent deposition of the mark at repressed genes.PLoS Genet. 2009 Jun;5(6):e1000506. doi: 10.1371/journal.pgen.1000506. Epub 2009 Jun 5. PLoS Genet. 2009. PMID: 19503595 Free PMC article.
-
Histone deacetylase inhibitors: a novel therapeutic approach to Huntington's disease (complex mechanism of neuronal death).Curr Alzheimer Res. 2006 Sep;3(4):403-8. doi: 10.2174/156720506778249407. Curr Alzheimer Res. 2006. PMID: 17017871 Review.
-
Drug targeting of dysregulated transcription in Huntington's disease.Prog Neurobiol. 2007 Nov;83(4):249-59. doi: 10.1016/j.pneurobio.2007.02.005. Epub 2007 Feb 23. Prog Neurobiol. 2007. PMID: 17379386 Free PMC article. Review.
Cited by
-
Pharmacology of epigenetics in brain disorders.Br J Pharmacol. 2010 Jan 1;159(2):285-303. doi: 10.1111/j.1476-5381.2009.00526.x. Epub 2009 Dec 15. Br J Pharmacol. 2010. PMID: 20015091 Free PMC article. Review.
-
Multiple Aspects of Gene Dysregulation in Huntington's Disease.Front Neurol. 2013 Oct 23;4:127. doi: 10.3389/fneur.2013.00127. Front Neurol. 2013. PMID: 24167500 Free PMC article. Review.
-
Balancing histone methylation activities in psychiatric disorders.Trends Mol Med. 2011 Jul;17(7):372-9. doi: 10.1016/j.molmed.2011.02.003. Epub 2011 Mar 21. Trends Mol Med. 2011. PMID: 21429800 Free PMC article. Review.
-
Genomic landscape of transcriptional and epigenetic dysregulation in early onset polyglutamine disease.J Neurosci. 2013 Jun 19;33(25):10471-82. doi: 10.1523/JNEUROSCI.0670-13.2013. J Neurosci. 2013. PMID: 23785159 Free PMC article.
-
MeCP2: a novel Huntingtin interactor.Hum Mol Genet. 2014 Feb 15;23(4):1036-44. doi: 10.1093/hmg/ddt499. Epub 2013 Oct 8. Hum Mol Genet. 2014. PMID: 24105466 Free PMC article.
References
-
- Augood SJ, Faull RL, Emson PC. Dopamine D1 and D2 receptor gene expression in the striatum in Huntington's disease. Ann Neurol. 1997;42:215–221. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. - PubMed
-
- Cattaneo E, Conti L. Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J Neurosci Res. 1998;53:223–234. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
