Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo
- PMID: 18390749
- PMCID: PMC3074963
- DOI: 10.4049/jimmunol.180.8.5645
Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo
Abstract
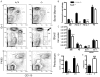
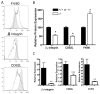
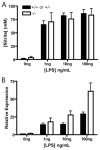
Several members of the Kruppel-like factor (KLF) family of transcription factors play important roles in differentiation, survival, and trafficking of blood and immune cell types. We demonstrate in this study that hematopoietic cells from KLF4(-/-) fetal livers (FL) contained normal numbers of functional hematopoietic progenitor cells, were radioprotective, and performed as well as KLF4(+/+) cells in competitive repopulation assays. However, hematopoietic "KLF4(-/-) chimeras" generated by transplantation of KLF4(-/-) fetal livers cells into lethally irradiated wild-type mice completely lacked circulating inflammatory (CD115(+)Gr1(+)) monocytes, and had reduced numbers of resident (CD115(+)Gr1(-)) monocytes. Although the numbers and function of peritoneal macrophages were normal in KLF4(-/-) chimeras, bone marrow monocytic cells from KLF4(-/-) chimeras expressed lower levels of key trafficking molecules and were more apoptotic. Thus, our in vivo loss-of-function studies demonstrate that KLF4, previously shown to mediate proinflammatory signaling in human macrophages in vitro, is essential for differentiation of mouse inflammatory monocytes, and is involved in the differentiation of resident monocytes. In addition, inducible expression of KLF4 in the HL60 human acute myeloid leukemia cell line stimulated monocytic differentiation and enhanced 12-O-tetradecanoylphorbol 13-acetate induced macrophage differentiation, but blocked all-trans-retinoic acid induced granulocytic differentiation of HL60 cells. The inflammation-selective effects of loss-of-KLF4 and the gain-of-KLF4-induced monocytic differentiation in HL60 cells identify KLF4 as a key regulator of monocytic differentiation and a potential target for translational immune modulation.
Figures
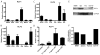

Similar articles
-
The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation.EMBO J. 2007 Sep 19;26(18):4138-48. doi: 10.1038/sj.emboj.7601824. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762869 Free PMC article.
-
Human skin dendritic cell fate is differentially regulated by the monocyte identity factor Kruppel-like factor 4 during steady state and inflammation.J Allergy Clin Immunol. 2017 Jun;139(6):1873-1884.e10. doi: 10.1016/j.jaci.2016.09.018. Epub 2016 Oct 11. J Allergy Clin Immunol. 2017. PMID: 27742396 Free PMC article.
-
A novel, myeloid transcription factor, C/EBP epsilon, is upregulated during granulocytic, but not monocytic, differentiation.Blood. 1997 Oct 1;90(7):2591-600. Blood. 1997. PMID: 9326225
-
Blood monocytes: development, heterogeneity, and relationship with dendritic cells.Annu Rev Immunol. 2009;27:669-92. doi: 10.1146/annurev.immunol.021908.132557. Annu Rev Immunol. 2009. PMID: 19132917 Review.
-
Regulation and consequences of monocytosis.Immunol Rev. 2014 Nov;262(1):167-78. doi: 10.1111/imr.12219. Immunol Rev. 2014. PMID: 25319334 Free PMC article. Review.
Cited by
-
KLF4 in Macrophages Attenuates TNFα-Mediated Kidney Injury and Fibrosis.J Am Soc Nephrol. 2019 Oct;30(10):1925-1938. doi: 10.1681/ASN.2019020111. Epub 2019 Jul 23. J Am Soc Nephrol. 2019. PMID: 31337692 Free PMC article.
-
The Role of KLF4 in Alzheimer's Disease.Front Cell Neurosci. 2018 Sep 21;12:325. doi: 10.3389/fncel.2018.00325. eCollection 2018. Front Cell Neurosci. 2018. PMID: 30297986 Free PMC article. Review.
-
Concise Review: Regulation of Self-Renewal in Normal and Malignant Hematopoietic Stem Cells by Krüppel-Like Factor 4.Stem Cells Transl Med. 2019 Jun;8(6):568-574. doi: 10.1002/sctm.18-0249. Epub 2019 Feb 21. Stem Cells Transl Med. 2019. PMID: 30790473 Free PMC article. Review.
-
TGF-β1 signaling and Krüppel-like factor 10 regulate bone marrow-derived proangiogenic cell differentiation, function, and neovascularization.Blood. 2011 Dec 8;118(24):6450-60. doi: 10.1182/blood-2011-06-363713. Epub 2011 Aug 9. Blood. 2011. PMID: 21828131 Free PMC article.
-
Transcriptional Control of Dendritic Cell Development.Annu Rev Immunol. 2016 May 20;34:93-119. doi: 10.1146/annurev-immunol-032713-120204. Epub 2015 Dec 23. Annu Rev Immunol. 2016. PMID: 26735697 Free PMC article. Review.
References
-
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. - PubMed
-
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–2448. - PubMed
-
- Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. - PubMed
-
- Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178:7632–7639. - PubMed
-
- Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

