Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein
- PMID: 18385239
- PMCID: PMC2395207
- DOI: 10.1128/JVI.02497-07
Identification of a highly conserved, functional nuclear localization signal within the N-terminal region of herpes simplex virus type 1 VP1-2 tegument protein
Abstract
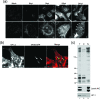
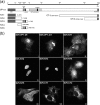
VP1-2 is a large structural protein assembled into the tegument compartment of the virion, conserved across the herpesviridae, and essential for virus replication. In herpes simplex virus (HSV) and pseudorabies virus, VP1-2 is tightly associated with the capsid. Studies of its assembly and function remain incomplete, although recent data indicate that in HSV, VP1-2 is recruited onto capsids in the nucleus, with this being required for subsequent recruitment of additional structural proteins. Here we have developed an antibody to characterize VP1-2 localization, observing the protein in both cytoplasmic and nuclear compartments, frequently in clusters in both locations. Within the nucleus, a subpopulation of VP1-2 colocalized with VP26 and VP5, though VP1-2-positive foci devoid of these components were observed. We note a highly conserved basic motif adjacent to the previously identified N-terminal ubiquitin hydrolase domain (DUB). The DUB domain in isolation exhibited no specific localization, but when extended to include the adjacent motif, it efficiently accumulated in the nucleus. Transfer of the isolated motif to a test protein, beta-galactosidase, conferred specific nuclear localization. Substitution of a single amino acid within the motif abolished the nuclear localization function. Deletion of the motif from intact VP1-2 abrogated its nuclear localization. Moreover, in a functional assay examining the ability of VP1-2 to complement growth of a VP1-2-ve mutant, deletion of the nuclear localization signal abolished complementation. The nuclear localization signal may be involved in transport of VP1-2 early in infection or to late assembly sites within the nucleus or, considering the potential existence of VP1-2 cleavage products, in selective localization of subdomains to different compartments.
Figures
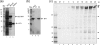

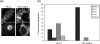

Similar articles
-
A Nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore.J Virol. 2012 Sep;86(17):8998-9014. doi: 10.1128/JVI.01209-12. Epub 2012 Jun 20. J Virol. 2012. PMID: 22718835 Free PMC article.
-
Functional analysis of nuclear localization signals in VP1-2 homologues from all herpesvirus subfamilies.J Virol. 2014 May;88(10):5391-405. doi: 10.1128/JVI.03797-13. Epub 2014 Feb 26. J Virol. 2014. PMID: 24574406 Free PMC article.
-
Characterization of the herpes simplex virus (HSV)-1 tegument protein VP1-2 during infection with the HSV temperature-sensitive mutant tsB7.J Gen Virol. 2009 Oct;90(Pt 10):2353-2363. doi: 10.1099/vir.0.012492-0. Epub 2009 Jul 8. J Gen Virol. 2009. PMID: 19587138
-
Identification of a minimal hydrophobic domain in the herpes simplex virus type 1 scaffolding protein which is required for interaction with the major capsid protein.J Virol. 1996 Jan;70(1):533-40. doi: 10.1128/JVI.70.1.533-540.1996. J Virol. 1996. PMID: 8523566 Free PMC article.
-
The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins.J Virol. 2007 Nov;81(21):11790-7. doi: 10.1128/JVI.01113-07. Epub 2007 Aug 22. J Virol. 2007. PMID: 17715218 Free PMC article.
Cited by
-
A Nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore.J Virol. 2012 Sep;86(17):8998-9014. doi: 10.1128/JVI.01209-12. Epub 2012 Jun 20. J Virol. 2012. PMID: 22718835 Free PMC article.
-
Tegument Proteins of Kaposi's Sarcoma-Associated Herpesvirus and Related Gamma-Herpesviruses.Front Microbiol. 2012 Mar 15;3:98. doi: 10.3389/fmicb.2012.00098. eCollection 2012. Front Microbiol. 2012. PMID: 22435068 Free PMC article.
-
Nuclear Cytoskeleton in Virus Infection.Int J Mol Sci. 2022 Jan 5;23(1):578. doi: 10.3390/ijms23010578. Int J Mol Sci. 2022. PMID: 35009004 Free PMC article. Review.
-
Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment.J Virol. 2009 Feb;83(4):1660-8. doi: 10.1128/JVI.01139-08. Epub 2008 Dec 10. J Virol. 2009. PMID: 19073727 Free PMC article.
-
Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1.J Virol. 2009 Jan;83(1):105-16. doi: 10.1128/JVI.01032-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971278 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

