Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum
- PMID: 18384229
- PMCID: PMC2276522
- DOI: 10.1371/journal.pmed.0050075
Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum
Abstract
Background: Tuberculous sputum provides a sample of bacilli that must be eliminated by chemotherapy and that may go on to transmit infection. A preliminary observation that Mycobacterium tuberculosis cells contain triacylglycerol lipid bodies in sputum, but not when growing in vitro, led us to investigate the extent of this phenomenon and its physiological basis.
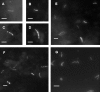
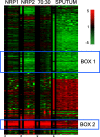
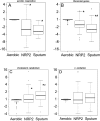
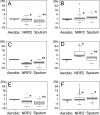
Methods and findings: Microscopy-positive sputum samples from the UK and The Gambia were investigated for their content of lipid body-positive mycobacteria by combined Nile red and auramine staining. All samples contained a lipid body-positive population varying from 3% to 86% of the acid-fast bacilli present. The recent finding that triacylglycerol synthase is expressed by mycobacteria when they enter in vitro nonreplicating persistence led us to investigate whether this state was also associated with lipid body formation. We found that, when placed in laboratory conditions inducing nonreplicating persistence, two M. tuberculosis strains had lipid body levels comparable to those found in sputum. We investigated these physiological findings further by comparing the M. tuberculosis transcriptome of growing and nonreplicating persistence cultures with that obtained directly from sputum samples. Although sputum has traditionally been thought to contain actively growing tubercle bacilli, our transcript analyses refute the hypothesis that these cells predominate. Rather, they reinforce the results of the lipid body analyses by revealing transcriptional signatures that can be clearly attributed to slowly replicating or nonreplicating mycobacteria. Finally, the lipid body count was highly correlated (R(2) = 0.64, p < 0.03) with time to positivity in diagnostic liquid cultures, thereby establishing a direct link between this cytological feature and the size of a potential nonreplicating population.
Conclusion: As nonreplicating tubercle bacilli are tolerant to the cidal action of antibiotics and resistant to multiple stresses, identification of this persister-like population of tubercle bacilli in sputum presents exciting and tractable new opportunities to investigate both responses to chemotherapy and the transmission of tuberculosis.
Conflict of interest statement
Figures
Similar articles
-
Association of slow recovery of Mycobacterium africanum-infected patients posttreatment with high content of Persister-Like bacilli in pretreatment sputum.Int J Mycobacteriol. 2016 Dec;5 Suppl 1:S99-S100. doi: 10.1016/j.ijmyco.2016.09.033. Epub 2016 Nov 11. Int J Mycobacteriol. 2016. PMID: 28043641
-
Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum.Microbiology (Reading). 2002 Oct;148(Pt 10):2951-8. doi: 10.1099/00221287-148-10-2951. Microbiology (Reading). 2002. PMID: 12368428
-
Automated identification of tubercle bacilli in sputum. A preliminary investigation.Anal Quant Cytol Histol. 1999 Aug;21(4):277-82. Anal Quant Cytol Histol. 1999. PMID: 10560504
-
Lipid droplets and the transcriptome of Mycobacterium tuberculosis from direct sputa: a literature review.Lipids Health Dis. 2021 Oct 3;20(1):129. doi: 10.1186/s12944-021-01550-5. Lipids Health Dis. 2021. PMID: 34602073 Free PMC article. Review.
-
Nonreplicating persistence of mycobacterium tuberculosis.Annu Rev Microbiol. 2001;55:139-63. doi: 10.1146/annurev.micro.55.1.139. Annu Rev Microbiol. 2001. PMID: 11544352 Review.
Cited by
-
Activities of drug combinations against Mycobacterium tuberculosis grown in aerobic and hypoxic acidic conditions.Antimicrob Agents Chemother. 2013 Mar;57(3):1428-33. doi: 10.1128/AAC.02154-12. Epub 2013 Jan 7. Antimicrob Agents Chemother. 2013. PMID: 23295931 Free PMC article.
-
Efficacy and safety of metronidazole for pulmonary multidrug-resistant tuberculosis.Antimicrob Agents Chemother. 2013 Aug;57(8):3903-9. doi: 10.1128/AAC.00753-13. Epub 2013 Jun 3. Antimicrob Agents Chemother. 2013. PMID: 23733467 Free PMC article. Clinical Trial.
-
Mycobacterium tuberculosis... Can we beat it? Report from a Euroscicon conference 2013.Virulence. 2013 Aug 15;4(6):499-503. doi: 10.4161/viru.25397. Epub 2013 Jun 18. Virulence. 2013. PMID: 23863609 Free PMC article. No abstract available.
-
Targeting multidrug-resistant tuberculosis (MDR-TB) by therapeutic vaccines.Med Microbiol Immunol. 2013 Apr;202(2):95-104. doi: 10.1007/s00430-012-0278-6. Epub 2012 Nov 10. Med Microbiol Immunol. 2013. PMID: 23143437 Review.
-
Crystal structure of reduced MsAcg, a putative nitroreductase from Mycobacterium smegmatis and a close homologue of Mycobacterium tuberculosis Acg.J Biol Chem. 2012 Dec 28;287(53):44372-83. doi: 10.1074/jbc.M112.406264. Epub 2012 Nov 12. J Biol Chem. 2012. PMID: 23148223 Free PMC article.
References
-
- WHO. Global tuberculosis control: surveillance, planning, financing. Geneva: WHO; 2007. WHO/HTM/TB/2007.376. Available: http://www.who.int/tb/publications/global_report/en/. Accessed 20 February 2008.
-
- Canetti G. The tubercle bacillus in the pulmonary lesion of man. Histobacteriology and its bearing on therapy of pulmonary tuberculosis. The tubercle bacillus in the pulmonary tuberculous lesion. New York: Springer Publishing Company; 1955. pp. 29–85.
-
- Young DB, Duncan K. Prospects for new interventions in the treatment and prevention of mycobacterial disease. Annu Rev Microbiol. 1995;49:641–673. - PubMed
-
- Mitchison DA. The search for new sterilizing anti-tuberculosis drugs. Front Biosci. 2004;9:1059–1072. - PubMed
-
- Garton NJ, Christensen H, Minnikin DE, Adegbola RA, Barer MR. Intracellular lipophilic inclusions of mycobacteria in vitro and in sputum. Microbiology. 2002;148:2951–2958. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

