An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer
- PMID: 18382681
- PMCID: PMC2270912
- DOI: 10.1371/journal.pone.0001908
An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer
Retraction in
-
Retraction: An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer.PLoS One. 2011;6(9):10.1371/annotation/8f94e479-4161-43a0-a28c-4c0460bb89a4. doi: 10.1371/annotation/8f94e479-4161-43a0-a28c-4c0460bb89a4. Epub 2011 Sep 2. PLoS One. 2011. PMID: 21912632 Free PMC article. No abstract available.
Abstract
Background: A major challenge in oncology is the selection of the most effective chemotherapeutic agents for individual patients, while the administration of ineffective chemotherapy increases mortality and decreases quality of life in cancer patients. This emphasizes the need to evaluate every patient's probability of responding to each chemotherapeutic agent and limiting the agents used to those most likely to be effective.
Methods and results: Using gene expression data on the NCI-60 and corresponding drug sensitivity, mRNA and microRNA profiles were developed representing sensitivity to individual chemotherapeutic agents. The mRNA signatures were tested in an independent cohort of 133 breast cancer patients treated with the TFAC (paclitaxel, 5-fluorouracil, adriamycin, and cyclophosphamide) chemotherapy regimen. To further dissect the biology of resistance, we applied signatures of oncogenic pathway activation and performed hierarchical clustering. We then used mRNA signatures of chemotherapy sensitivity to identify alternative therapeutics for patients resistant to TFAC. Profiles from mRNA and microRNA expression data represent distinct biologic mechanisms of resistance to common cytotoxic agents. The individual mRNA signatures were validated in an independent dataset of breast tumors (P = 0.002, NPV = 82%). When the accuracy of the signatures was analyzed based on molecular variables, the predictive ability was found to be greater in basal-like than non basal-like patients (P = 0.03 and P = 0.06). Samples from patients with co-activated Myc and E2F represented the cohort with the lowest percentage (8%) of responders. Using mRNA signatures of sensitivity to other cytotoxic agents, we predict that TFAC non-responders are more likely to be sensitive to docetaxel (P = 0.04), representing a viable alternative therapy.
Conclusions: Our results suggest that the optimal strategy for chemotherapy sensitivity prediction integrates molecular variables such as ER and HER2 status with corresponding microRNA and mRNA expression profiles. Importantly, we also present evidence to support the concept that analysis of molecular variables can present a rational strategy to identifying alternative therapeutic opportunities.
Conflict of interest statement
Figures
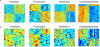


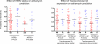

Comment in
-
Findings of research misconduct.NIH Guide Grants Contracts. 2015 Nov 20:NOT-OD-16-021. NIH Guide Grants Contracts. 2015. PMID: 26601329 Free PMC article. No abstract available.
Similar articles
-
Inhibitory effects of combinations of HER-2/neu antibody and chemotherapeutic agents used for treatment of human breast cancers.Oncogene. 1999 Apr 1;18(13):2241-51. doi: 10.1038/sj.onc.1202526. Oncogene. 1999. PMID: 10327070
-
Breast cancer--response rates to chemotherapeutic agents studied in vitro.Anticancer Res. 2003 Jul-Aug;23(4):3405-11. Anticancer Res. 2003. PMID: 12926081
-
miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer.Cell Death Dis. 2016 Jun 30;7(6):e2291. doi: 10.1038/cddis.2016.194. Cell Death Dis. 2016. PMID: 27362808 Free PMC article.
-
Docetaxel: an update of its use in advanced breast cancer.Drugs. 2000 Mar;59(3):621-51. doi: 10.2165/00003495-200059030-00015. Drugs. 2000. PMID: 10776837 Review.
-
Emerging role of taxanes in adjuvant and neoadjuvant therapy for breast cancer: the potential and the questions.Surg Clin North Am. 2003 Aug;83(4):943-71. doi: 10.1016/S0039-6109(03)00071-9. Surg Clin North Am. 2003. PMID: 12875604 Review.
Cited by
-
Discovery of IL-18 as a novel secreted protein contributing to doxorubicin resistance by comparative secretome analysis of MCF-7 and MCF-7/Dox.PLoS One. 2011;6(9):e24684. doi: 10.1371/journal.pone.0024684. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931812 Free PMC article.
-
The use of genomic information to optimize cancer chemotherapy.Semin Oncol. 2011 Apr;38(2):186-95. doi: 10.1053/j.seminoncol.2011.01.005. Semin Oncol. 2011. PMID: 21421109 Free PMC article. Review.
-
[Advances in tumor chemo-resistance regulated by MicroRNA].Zhongguo Fei Ai Za Zhi. 2014 Oct 20;17(10):741-9. doi: 10.3779/j.issn.1009-3419.2014.10.06. Zhongguo Fei Ai Za Zhi. 2014. PMID: 25342041 Free PMC article. Review. Chinese.
-
Micro-RNAs as clinical biomarkers and therapeutic targets in breast cancer: Quo vadis?World J Clin Oncol. 2014 May 10;5(2):71-81. doi: 10.5306/wjco.v5.i2.71. World J Clin Oncol. 2014. PMID: 24829853 Free PMC article. Review.
-
Key signalling nodes in mammary gland development and cancer: Myc.Breast Cancer Res. 2009;11(5):210. doi: 10.1186/bcr2406. Breast Cancer Res. 2009. PMID: 19849814 Free PMC article. Review.
References
-
- Herbst RS, Bajorin DF, Bleiberg H, Blum D, Hao D, et al. Clinical Cancer Advances 2005: major research advances in cancer treatment, prevention, and screening–a report from the American Society of Clinical Oncology. J Clin Oncol. 2006;24:190–205. - PubMed
-
- Breathnach OS, Freidlin B, Conley B, Green MR, Johnson DH, et al. Twenty-two years of phase III trials for patients with advanced non-small-cell lung cancer: sobering results. J Clin Oncol. 2001;19:1734–1742. - PubMed
-
- Potti A, Dressman HK, Bild A, Riedel RF, Chan G, et al. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12:1294–1300. - PubMed
-
- Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet. 2007;8:601–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

