Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase
- PMID: 18378697
- PMCID: PMC2423282
- DOI: 10.1128/MCB.00087-08
Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase
Abstract
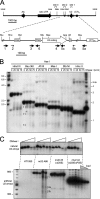
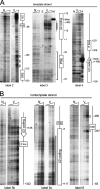
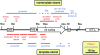
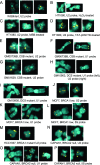
In mammals, small multigene families generate spliceosomal U snRNAs that are nearly as abundant as rRNA. Using the tandemly repeated human U2 genes as a model, we show by footprinting with DNase I and permanganate that nearly all sequences between the enhancer-like distal sequence element and the initiation site are protected during interphase whereas the upstream half of the U2 snRNA coding region is exposed. We also show by chromatin immunoprecipitation that the SNAPc complex, which binds the TATA-like proximal sequence element, is removed at metaphase but remains bound under conditions that induce locus-specific metaphase fragility of the U2 genes, such as loss of CSB, BRCA1, or BRCA2 function, treatment with actinomycin D, or overexpression of the tetrameric p53 C terminus. We propose that the U2 snRNA promoter establishes a persistently open state to facilitate rapid reinitiation and perhaps also to bypass TFIIH-dependent promoter melting; this open state would then be disassembled to allow metaphase chromatin condensation.
Figures


Similar articles
-
Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes.Mol Cell. 2000 May;5(5):801-10. doi: 10.1016/s1097-2765(00)80320-2. Mol Cell. 2000. PMID: 10882116
-
A tandem array of minimal U1 small nuclear RNA genes is sufficient to generate a new adenovirus type 12-inducible chromosome fragile site.J Virol. 1998 May;72(5):4205-11. doi: 10.1128/JVI.72.5.4205-4211.1998. J Virol. 1998. PMID: 9557709 Free PMC article.
-
Metaphase fragility of the human RNU1 and RNU2 loci is induced by actinomycin D through a p53-dependent pathway.Hum Mol Genet. 1998 Apr;7(4):609-17. doi: 10.1093/hmg/7.4.609. Hum Mol Genet. 1998. PMID: 9499413
-
Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc).Crit Rev Biochem Mol Biol. 2011 Feb;46(1):11-26. doi: 10.3109/10409238.2010.518136. Epub 2010 Oct 6. Crit Rev Biochem Mol Biol. 2011. PMID: 20925482 Review.
-
Expression of human snRNA genes from beginning to end.Biochem Soc Trans. 2008 Aug;36(Pt 4):590-4. doi: 10.1042/BST0360590. Biochem Soc Trans. 2008. PMID: 18631122 Review.
Cited by
-
Functional analysis of the integrator subunit 12 identifies a microdomain that mediates activation of the Drosophila integrator complex.J Biol Chem. 2013 Feb 15;288(7):4867-77. doi: 10.1074/jbc.M112.425892. Epub 2013 Jan 3. J Biol Chem. 2013. PMID: 23288851 Free PMC article.
-
Interaction between the Cockayne syndrome B and p53 proteins: implications for aging.Aging (Albany NY). 2012 Feb;4(2):89-97. doi: 10.18632/aging.100439. Aging (Albany NY). 2012. PMID: 22383384 Free PMC article. Review.
-
Human snRNA genes use polyadenylation factors to promote efficient transcription termination.Nucleic Acids Res. 2014 Jan;42(1):264-75. doi: 10.1093/nar/gkt892. Epub 2013 Oct 4. Nucleic Acids Res. 2014. PMID: 24097444 Free PMC article.
-
The CSB chromatin remodeler regulates PARP1- and PARP2-mediated single-strand break repair at actively transcribed DNA regions.Nucleic Acids Res. 2023 Aug 11;51(14):7342-7356. doi: 10.1093/nar/gkad515. Nucleic Acids Res. 2023. PMID: 37326017 Free PMC article.
-
Merging Established Mechanisms with New Insights: Condensates, Hubs, and the Regulation of RNA Polymerase II Transcription.J Mol Biol. 2022 Jan 15;434(1):167216. doi: 10.1016/j.jmb.2021.167216. Epub 2021 Aug 30. J Mol Biol. 2022. PMID: 34474085 Free PMC article. Review.
References
-
- Akman, S. A., J. H. Doroshow, and M. Dizdaroglu. 1990. Base modifications in plasmid DNA caused by potassium permanganate. Arch. Biochem. Biophys. 282202-205. - PubMed
-
- Ares, M., Jr., J. S. Chung, L. Giglio, and A. M. Weiner. 1987. Distinct factors with Sp1 and NF-A specificities bind to adjacent functional elements of the human U2 snRNA gene enhancer. Genes Dev. 1808-817. - PubMed
-
- Baillat, D., M. A. Hakimi, A. M. Naar, A. Shilatifard, N. Cooch, and R. Shiekhattar. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123265-276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
