Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies
- PMID: 18367544
- PMCID: PMC2397305
- DOI: 10.1091/mbc.e07-12-1259
Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies
Abstract
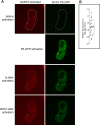
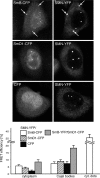
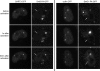
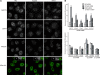
The Cajal body (CB) is a nuclear structure closely associated with import and biogenesis of small nuclear ribonucleoprotein particles (snRNPs). Here, we tested whether CBs also contain mature snRNPs and whether CB integrity depends on the ongoing snRNP splicing cycle. Sm proteins tagged with photoactivatable and color-maturing variants of fluorescent proteins were used to monitor snRNP behavior in living cells over time; mature snRNPs accumulated in CBs, traveled from one CB to another, and they were not preferentially replaced by newly imported snRNPs. To test whether CB integrity depends on the snRNP splicing cycle, two human orthologues of yeast proteins involved in distinct steps in spliceosome disassembly after splicing, hPrp22 and hNtr1, were depleted by small interfering RNA treatment. Surprisingly, depletion of either protein led to the accumulation of U4/U6 snRNPs in CBs, suggesting that reassembly of the U4/U6.U5 tri-snRNP was delayed. Accordingly, a relative decrease in U5 snRNPs compared with U4/U6 snRNPs was observed in CBs, as well as in nuclear extracts of treated cells. Together, the data show that particular phases of the spliceosome cycle are compartmentalized in living cells, with reassembly of the tri-snRNP occurring in CBs.
Figures
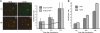
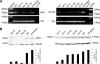


Comment in
- Mol Biol Cell. 19:2349.
Similar articles
-
In vivo kinetics of U4/U6·U5 tri-snRNP formation in Cajal bodies.Mol Biol Cell. 2011 Feb 15;22(4):513-23. doi: 10.1091/mbc.E10-07-0560. Epub 2010 Dec 22. Mol Biol Cell. 2011. PMID: 21177826 Free PMC article.
-
RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies.EMBO J. 2004 Aug 4;23(15):3000-9. doi: 10.1038/sj.emboj.7600296. Epub 2004 Jul 15. EMBO J. 2004. PMID: 15257298 Free PMC article.
-
Interaction of the human autoantigen p150 with splicing snRNPs.J Cell Sci. 1993 Jul;105 ( Pt 3):685-97. doi: 10.1242/jcs.105.3.685. J Cell Sci. 1993. PMID: 8408296
-
The assembly of a spliceosomal small nuclear ribonucleoprotein particle.Nucleic Acids Res. 2008 Nov;36(20):6482-93. doi: 10.1093/nar/gkn658. Epub 2008 Oct 14. Nucleic Acids Res. 2008. PMID: 18854356 Free PMC article. Review.
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
Cited by
-
The Cajal body and histone locus body.Cold Spring Harb Perspect Biol. 2010 Jul;2(7):a000653. doi: 10.1101/cshperspect.a000653. Epub 2010 May 26. Cold Spring Harb Perspect Biol. 2010. PMID: 20504965 Free PMC article. Review.
-
The life of U6 small nuclear RNA, from cradle to grave.RNA. 2018 Apr;24(4):437-460. doi: 10.1261/rna.065136.117. Epub 2018 Jan 24. RNA. 2018. PMID: 29367453 Free PMC article. Review.
-
Evolutionary Analysis of the Mammalian Tuftelin Sequence Reveals Features of Functional Importance.J Mol Evol. 2017 Apr;84(4):214-224. doi: 10.1007/s00239-017-9789-5. Epub 2017 Apr 13. J Mol Evol. 2017. PMID: 28409196
-
Pathogenic Variants in USH1G/SANS Alter Protein Interaction with Pre-RNA Processing Factors PRPF6 and PRPF31 of the Spliceosome.Int J Mol Sci. 2023 Dec 18;24(24):17608. doi: 10.3390/ijms242417608. Int J Mol Sci. 2023. PMID: 38139438 Free PMC article.
-
In vivo kinetics of U4/U6·U5 tri-snRNP formation in Cajal bodies.Mol Biol Cell. 2011 Feb 15;22(4):513-23. doi: 10.1091/mbc.E10-07-0560. Epub 2010 Dec 22. Mol Biol Cell. 2011. PMID: 21177826 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

