A sequence-based survey of the complex structural organization of tumor genomes
- PMID: 18364049
- PMCID: PMC2397511
- DOI: 10.1186/gb-2008-9-3-r59
A sequence-based survey of the complex structural organization of tumor genomes
Abstract
Background: The genomes of many epithelial tumors exhibit extensive chromosomal rearrangements. All classes of genome rearrangements can be identified using end sequencing profiling, which relies on paired-end sequencing of cloned tumor genomes.
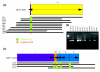
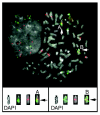
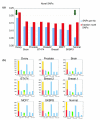
Results: In the present study brain, breast, ovary, and prostate tumors, along with three breast cancer cell lines, were surveyed using end sequencing profiling, yielding the largest available collection of sequence-ready tumor genome breakpoints and providing evidence that some rearrangements may be recurrent. Sequencing and fluorescence in situ hybridization confirmed translocations and complex tumor genome structures that include co-amplification and packaging of disparate genomic loci with associated molecular heterogeneity. Comparison of the tumor genomes suggests recurrent rearrangements. Some are likely to be novel structural polymorphisms, whereas others may be bona fide somatic rearrangements. A recurrent fusion transcript in breast tumors and a constitutional fusion transcript resulting from a segmental duplication were identified. Analysis of end sequences for single nucleotide polymorphisms revealed candidate somatic mutations and an elevated rate of novel single nucleotide polymorphisms in an ovarian tumor.
Conclusion: These results suggest that the genomes of many epithelial tumors may be far more dynamic and complex than was previously appreciated and that genomic fusions, including fusion transcripts and proteins, may be common, possibly yielding tumor-specific biomarkers and therapeutic targets.
Figures



Similar articles
-
A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome.Genome Res. 2009 Feb;19(2):167-77. doi: 10.1101/gr.080259.108. Epub 2008 Dec 3. Genome Res. 2009. PMID: 19056696 Free PMC article.
-
Decoding the fine-scale structure of a breast cancer genome and transcriptome.Genome Res. 2006 Mar;16(3):394-404. doi: 10.1101/gr.4247306. Epub 2006 Feb 3. Genome Res. 2006. PMID: 16461635 Free PMC article.
-
Transcriptional consequences of genomic structural aberrations in breast cancer.Genome Res. 2011 May;21(5):676-87. doi: 10.1101/gr.113225.110. Epub 2011 Apr 5. Genome Res. 2011. PMID: 21467264 Free PMC article.
-
Characterising chromosome rearrangements: recent technical advances in molecular cytogenetics.Heredity (Edinb). 2012 Jan;108(1):75-85. doi: 10.1038/hdy.2011.100. Epub 2011 Nov 16. Heredity (Edinb). 2012. PMID: 22086080 Free PMC article. Review.
-
Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes.PLoS Genet. 2005 Dec;1(6):e49. doi: 10.1371/journal.pgen.0010049. PLoS Genet. 2005. PMID: 16444292 Free PMC article. Review.
Cited by
-
Optimizing PCR assays for DNA-based cancer diagnostics.J Comput Biol. 2010 Mar;17(3):369-81. doi: 10.1089/cmb.2009.0203. J Comput Biol. 2010. PMID: 20377451 Free PMC article.
-
Analysis of next-generation genomic data in cancer: accomplishments and challenges.Hum Mol Genet. 2010 Oct 15;19(R2):R188-96. doi: 10.1093/hmg/ddq391. Epub 2010 Sep 15. Hum Mol Genet. 2010. PMID: 20843826 Free PMC article. Review.
-
PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data.Genome Biol. 2009 Feb 23;10(2):R23. doi: 10.1186/gb-2009-10-2-r23. Genome Biol. 2009. PMID: 19236709 Free PMC article.
-
A sequence-level map of chromosomal breakpoints in the MCF-7 breast cancer cell line yields insights into the evolution of a cancer genome.Genome Res. 2009 Feb;19(2):167-77. doi: 10.1101/gr.080259.108. Epub 2008 Dec 3. Genome Res. 2009. PMID: 19056696 Free PMC article.
-
A geometric approach for classification and comparison of structural variants.Bioinformatics. 2009 Jun 15;25(12):i222-30. doi: 10.1093/bioinformatics/btp208. Bioinformatics. 2009. PMID: 19477992 Free PMC article.
References
-
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. - DOI - PubMed
-
- Raphael BJ, Volik S, Collins C, Pevzner PA. Reconstructing tumor genome architectures. Bioinformatics. 2003;19(suppl 2):II162–II171. - PubMed
Publication types
MeSH terms
Grants and funding
- U24 CA 126551/CA/NCI NIH HHS/United States
- U01HL66728/HL/NHLBI NIH HHS/United States
- U24 CA 126477/CA/NCI NIH HHS/United States
- R01 GM057070-10/GM/NIGMS NIH HHS/United States
- U54 CA 112970/CA/NCI NIH HHS/United States
- P30 CA 82103/CA/NCI NIH HHS/United States
- P50 CA 58207/CA/NCI NIH HHS/United States
- CA5807/CA/NCI NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 GM057070/GM/NIGMS NIH HHS/United States
- P50 CA 83639/CA/NCI NIH HHS/United States
- P01 CA 64602/CA/NCI NIH HHS/United States
- P50 CA69568/CA/NCI NIH HHS/United States
- GM00806-06/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

