An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor
- PMID: 18362181
- PMCID: PMC2373575
- DOI: 10.1083/jcb.200708115
An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor
Abstract
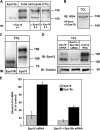
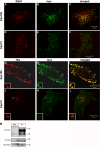
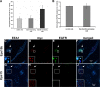
Down-regulation of activated and ubiquitinated growth factor (GF) receptors by endocytosis and subsequent lysosomal degradation ensures attenuation of GF signaling. The ubiquitin-binding adaptor protein Eps15 (epidermal growth factor receptor [EGFR] pathway substrate 15) functions in endocytosis of such receptors. Here, we identify an Eps15 isoform, Eps15b, and demonstrate its expression in human cells and conservation across vertebrate species. Although both Eps15 and Eps15b interact with the endosomal sorting protein Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) in vitro, we find that Hrs specifically binds Eps15b in vivo (whereas adaptor protein 2 preferentially interacts with Eps15). Although Eps15 mainly localizes to clathrin-coated pits at the plasma membrane, Eps15b localizes to Hrs-positive microdomains on endosomes. Eps15b overexpression, similarly to Hrs overexpression, inhibits ligand-mediated degradation of EGFR, whereas Eps15 is without effect. Similarly, depletion of Eps15b but not Eps15 delays degradation and promotes recycling of EGFR. These results indicate that Eps15b is an endosomally localized isoform of Eps15 that is present in the Hrs complex via direct Hrs interaction and important for the sorting function of this complex.
Figures



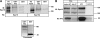


Similar articles
-
STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes.J Biol Chem. 2003 Apr 4;278(14):12513-21. doi: 10.1074/jbc.M210843200. Epub 2003 Jan 27. J Biol Chem. 2003. PMID: 12551915
-
Eps15 is recruited to the plasma membrane upon epidermal growth factor receptor activation and localizes to components of the endocytic pathway during receptor internalization.Mol Biol Cell. 1999 Feb;10(2):417-34. doi: 10.1091/mbc.10.2.417. Mol Biol Cell. 1999. PMID: 9950686 Free PMC article.
-
Recycling of the epidermal growth factor receptor is mediated by a novel form of the clathrin adaptor protein Eps15.J Biol Chem. 2011 Oct 7;286(40):35196-208. doi: 10.1074/jbc.M111.247577. Epub 2011 Aug 8. J Biol Chem. 2011. PMID: 21832070 Free PMC article.
-
Hrs and endocytic sorting of ubiquitinated membrane proteins.Cell Struct Funct. 2002 Dec;27(6):403-8. doi: 10.1247/csf.27.403. Cell Struct Funct. 2002. PMID: 12576633 Review.
-
Epidermal growth factor pathway substrate 15, Eps15.Int J Biochem Cell Biol. 1999 Aug;31(8):805-9. doi: 10.1016/s1357-2725(99)00042-4. Int J Biochem Cell Biol. 1999. PMID: 10481267 Review.
Cited by
-
Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane.Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):11857-62. doi: 10.1073/pnas.1302918110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818590 Free PMC article.
-
The Role of NEDD4 E3 Ubiquitin-Protein Ligases in Parkinson's Disease.Genes (Basel). 2022 Mar 14;13(3):513. doi: 10.3390/genes13030513. Genes (Basel). 2022. PMID: 35328067 Free PMC article. Review.
-
Motor and Sensory Deficits in the teetering Mice Result from Mutation of the ESCRT Component HGS.PLoS Genet. 2015 Jun 26;11(6):e1005290. doi: 10.1371/journal.pgen.1005290. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26115514 Free PMC article.
-
Neutral sphingomyelinase 2 controls exosome secretion by counteracting V-ATPase-mediated endosome acidification.J Cell Sci. 2022 Mar 1;135(5):jcs259324. doi: 10.1242/jcs.259324. Epub 2022 Feb 28. J Cell Sci. 2022. PMID: 35050379 Free PMC article.
-
SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling.Mol Biol Cell. 2009 Mar;20(6):1816-32. doi: 10.1091/mbc.e08-09-0894. Epub 2009 Jan 21. Mol Biol Cell. 2009. PMID: 19158374 Free PMC article.
References
-
- Babst, M., D.J. Katzmann, E.J. Estepa-Sabal, T. Meerloo, and S.D. Emr. 2002. a. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. - PubMed
-
- Babst, M., D.J. Katzmann, W.B. Snyder, B. Wendland, and S.D. Emr. 2002. b. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell. 3:283–289. - PubMed
-
- Bache, K.G., C. Raiborg, A. Mehlum, and H. Stenmark. 2003. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J. Biol. Chem. 278:12513–12521. - PubMed
-
- Bean, A.J., S. Davanger, M.F. Chou, B. Gerhardt, S. Tsujimoto, and Y. Chang. 2000. Hrs-2 regulates receptor-mediated endocytosis via interactions with Eps15. J. Biol. Chem. 275:15271–15278. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

