Optimization of RNA extraction from FFPE tissues for expression profiling in the DASL assay
- PMID: 18361796
- PMCID: PMC2672087
- DOI: 10.2144/000112703
Optimization of RNA extraction from FFPE tissues for expression profiling in the DASL assay
Abstract
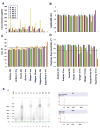
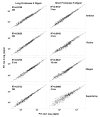
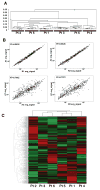
Formalin-fixed paraffin-embedded (FFPE) breast tumor tissues are readily available and represent a largely untapped, vast resource for molecular profiling of clinical samples with long-term follow-up data. We have optimized the conditions and parameters that result in the preparation of total RNA that is of the necessary quality for use in the DASL (cDNA-mediated annealing, selection, extension, and ligation) assay in which expression of 502 genes are analyzed simultaneously using as little as 100 ng of input RNA.
Conflict of interest statement
Figures
Similar articles
-
Reliable gene expression profiling of formalin-fixed paraffin-embedded breast cancer tissue (FFPE) using cDNA-mediated annealing, extension, selection, and ligation whole-genome (DASL WG) assay.BMC Med Genomics. 2016 Aug 20;9(1):54. doi: 10.1186/s12920-016-0215-4. BMC Med Genomics. 2016. PMID: 27542606 Free PMC article.
-
Gene expression profiles from formalin fixed paraffin embedded breast cancer tissue are largely comparable to fresh frozen matched tissue.PLoS One. 2011 Feb 11;6(2):e17163. doi: 10.1371/journal.pone.0017163. PLoS One. 2011. PMID: 21347257 Free PMC article.
-
Gene expression profiling of formalin-fixed, paraffin-embedded familial breast tumours using the whole genome-DASL assay.J Pathol. 2010 Aug;221(4):452-61. doi: 10.1002/path.2728. J Pathol. 2010. PMID: 20593485
-
Tissue handling and specimen preparation in surgical pathology: issues concerning the recovery of nucleic acids from formalin-fixed, paraffin-embedded tissue.Arch Pathol Lab Med. 2008 Dec;132(12):1929-35. doi: 10.5858/132.12.1929. Arch Pathol Lab Med. 2008. PMID: 19061293 Review.
-
Fusion transcript discovery using RNA sequencing in formalin-fixed paraffin-embedded specimen.Crit Rev Oncol Hematol. 2021 Apr;160:103303. doi: 10.1016/j.critrevonc.2021.103303. Epub 2021 Mar 20. Crit Rev Oncol Hematol. 2021. PMID: 33757837 Review.
Cited by
-
Simultaneous recovery of DNA and RNA from formalin-fixed paraffin-embedded tissue and application in epidemiologic studies.Cancer Epidemiol Biomarkers Prev. 2010 Apr;19(4):973-7. doi: 10.1158/1055-9965.EPI-10-0091. Epub 2010 Mar 23. Cancer Epidemiol Biomarkers Prev. 2010. Retraction in: Cancer Epidemiol Biomarkers Prev. 2014 Jun;23(6):1132. doi: 10.1158/1055-9965.EPI-14-0395. PMID: 20332269 Free PMC article. Retracted.
-
Gene expression array testing of FFPE archival breast tumor samples: an optimized protocol for WG-DASL sample preparation.Breast Cancer Res Treat. 2011 Feb;125(3):879-83. doi: 10.1007/s10549-010-1159-6. Epub 2010 Sep 15. Breast Cancer Res Treat. 2011. PMID: 20842525 Free PMC article.
-
Akt inhibitors MK-2206 and nelfinavir overcome mTOR inhibitor resistance in diffuse large B-cell lymphoma.Clin Cancer Res. 2012 May 1;18(9):2534-44. doi: 10.1158/1078-0432.CCR-11-1407. Epub 2012 Feb 14. Clin Cancer Res. 2012. PMID: 22338016 Free PMC article.
-
Protein-coding and microRNA biomarkers of recurrence of prostate cancer following radical prostatectomy.Am J Pathol. 2011 Jul;179(1):46-54. doi: 10.1016/j.ajpath.2011.03.008. Epub 2011 May 3. Am J Pathol. 2011. PMID: 21703393 Free PMC article.
-
Global transcriptome analysis of formalin-fixed prostate cancer specimens identifies biomarkers of disease recurrence.Cancer Res. 2014 Jun 15;74(12):3228-37. doi: 10.1158/0008-5472.CAN-13-2699. Epub 2014 Apr 8. Cancer Res. 2014. PMID: 24713434 Free PMC article.
References
-
- Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006;15:229–236. - PubMed
-
- Rupp GM, Locker J. Purification and analysis of RNA from paraffin-embedded tissues. Biotechniques. 1988;6:56–60. - PubMed
-
- Penland SK, Keku TO, Torrice C, He X, Krishnamurthy J, Hoadley KA, Woosley JT, Thomas NE, et al. RNA expression analysis of formalin-fixed paraffin-embedded tumors. Lab Invest. 2007;87:383–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
