Elucidating the germination transcriptional program using small molecules
- PMID: 18359847
- PMCID: PMC2330302
- DOI: 10.1104/pp.107.110841
Elucidating the germination transcriptional program using small molecules
Abstract
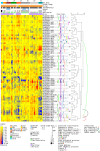
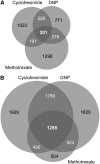
The transition from seed to seedling is mediated by germination, a complex process that starts with imbibition and completes with radicle emergence. To gain insight into the transcriptional program mediating germination, previous studies have compared the transcript profiles of dry, dormant, and germinating after-ripened Arabidopsis (Arabidopsis thaliana) seeds. While informative, these approaches did not distinguish the transcriptional responses due to imbibition, shifts in metabolism, or breaking of dormancy from those triggered by the initiation of germination. In this study, three mechanistically distinct small molecules that inhibit Arabidopsis seed germination (methotrexate, 2, 4-dinitrophenol, and cycloheximide) were identified using a small-molecule screen and used to probe the germination transcriptome. Germination-responsive transcripts were defined as those with significantly altered transcript abundance across all inhibitory treatments with respect to control germinating seeds, using data from ATH1 microarrays. This analysis identified numerous germination regulators as germination responsive, including the DELLA proteins GAI, RGA, and RGL3, the abscisic acid-insensitive proteins ABI4, ABI5, ABI8, and FRY1, and the gibberellin receptor GID1A. To help visualize these and other publicly available seed microarray data, we designed a seed mRNA expression browser using the electronic Fluorescent Pictograph platform. An overall decrease in gene expression and a 5-fold greater number of transcripts identified as statistically down-regulated in drug-inhibited seeds point to a role for mRNA degradation or turnover during seed germination. The genes identified in our study as responsive to germination define potential uncharacterized regulators of this process and provide a refined transcriptional signature for germinating Arabidopsis seeds.
Figures



Similar articles
-
Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis.Plant Physiol. 2004 Jun;135(2):1008-19. doi: 10.1104/pp.104.039578. Epub 2004 Jun 1. Plant Physiol. 2004. PMID: 15173565 Free PMC article.
-
Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition.Plant Mol Biol. 2010 May;73(1-2):119-29. doi: 10.1007/s11103-010-9603-x. Epub 2010 Jan 23. Plant Mol Biol. 2010. PMID: 20099072
-
Loss of Arabidopsis thaliana Seed Dormancy is Associated with Increased Accumulation of the GID1 GA Hormone Receptors.Plant Cell Physiol. 2015 Sep;56(9):1773-85. doi: 10.1093/pcp/pcv084. Epub 2015 Jul 1. Plant Cell Physiol. 2015. PMID: 26136598
-
First off the mark: early seed germination.J Exp Bot. 2011 Jun;62(10):3289-309. doi: 10.1093/jxb/err030. Epub 2011 Mar 23. J Exp Bot. 2011. PMID: 21430292 Review.
-
Post-genomics dissection of seed dormancy and germination.Trends Plant Sci. 2008 Jan;13(1):7-13. doi: 10.1016/j.tplants.2007.11.002. Epub 2007 Dec 21. Trends Plant Sci. 2008. PMID: 18160329 Review.
Cited by
-
The ARC1 E3 ligase gene is frequently deleted in self-compatible Brassicaceae species and has a conserved role in Arabidopsis lyrata self-pollen rejection.Plant Cell. 2012 Nov;24(11):4607-20. doi: 10.1105/tpc.112.104943. Epub 2012 Nov 30. Plant Cell. 2012. PMID: 23204404 Free PMC article.
-
Proteomic Dissection of Seed Germination and Seedling Establishment in Brassica napus.Front Plant Sci. 2016 Oct 24;7:1482. doi: 10.3389/fpls.2016.01482. eCollection 2016. Front Plant Sci. 2016. PMID: 27822216 Free PMC article.
-
Integration of proteomic and genomic approaches to dissect seed germination vigor in Brassica napus seeds differing in oil content.BMC Plant Biol. 2019 Jan 11;19(1):21. doi: 10.1186/s12870-018-1624-7. BMC Plant Biol. 2019. PMID: 30634904 Free PMC article.
-
A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates.PLoS Pathog. 2008 Nov;4(11):e1000214. doi: 10.1371/journal.ppat.1000214. Epub 2008 Nov 21. PLoS Pathog. 2008. PMID: 19023412 Free PMC article.
-
Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins.Science. 2009 May 22;324(5930):1068-71. doi: 10.1126/science.1173041. Epub 2009 Apr 30. Science. 2009. PMID: 19407142 Free PMC article.
References
-
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN (2005) Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ 28 500–512 - PubMed
-
- Bewley JD (1997) Breaking down the walls—a role for endo-beta-mannanase in release from seed dormancy? Trends Plant Sci 2 464–469
-
- Bewley JD, Black M (1994) Seeds: Physiology of Development and Germination, Ed 2. Plenum Press, New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

