Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter
- PMID: 18353421
- PMCID: PMC2632587
- DOI: 10.1016/j.virol.2008.02.021
Early growth response-1 protein is induced by JC virus infection and binds and regulates the JC virus promoter
Abstract
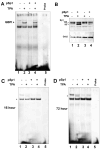
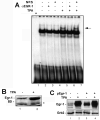
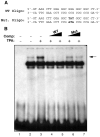
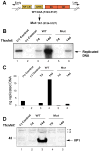
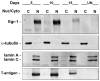
JC virus (JCV) is a human polyomavirus that can emerge from a latent state to cause the cytolytic destruction of oligodendrocytes in the brain resulting in the fatal demyelinating disease, progressive multifocal leukoencephalopathy (PML). Previous studies described a cis-acting transcriptional regulatory element in the JCV non-coding control region (NCCR) that is involved in the response of JCV to cytokines. This consists of a 23 base pair GGA/C rich sequence (GRS) near the replication origin (5112 to +4) that contains potential binding sites for Sp1 and Egr-1. Gel shift analysis showed that Egr-1, but not Sp1, bound to GRS. Evidence is presented that the GRS gel shift seen on cellular stimulation is due to Egr-1. Thus, TPA-induced GRS gel shift could be blocked by antibody to Egr-1. Further, the TPA-induced GRS DNA/protein complex was isolated and found to contain Egr-1 by Western blot. No other Egr-1 sites were found in the JCV NCCR. Functionally, Egr-1 was found to stimulate transcription of JCV late promoter but not early promoter reporter constructs. Mutation of the Egr-1 site abrogated Egr-1 binding and virus with the mutated Egr-1 site showed markedly reduced VP1 expression and DNA replication. Infection of primary astrocytes by wild-type JCV induced Egr-1 nuclear expression that was maximal at 5-10 days post-infection. Finally, upregulation of Egr-1 was detected in PML by immunohistochemistry. These data suggest that Egr-1 induction may be important in the life cycle of JCV and PML pathogenesis.
Figures



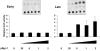

Similar articles
-
Evidence for involvement of transforming growth factor beta1 signaling pathway in activation of JC virus in human immunodeficiency virus 1-associated progressive multifocal leukoencephalopathy.Arch Pathol Lab Med. 2004 Mar;128(3):282-91. doi: 10.5858/2004-128-282-EFIOTG. Arch Pathol Lab Med. 2004. PMID: 14987161
-
Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate.J Virol. 2010 Oct;84(20):10448-56. doi: 10.1128/JVI.00614-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686041 Free PMC article.
-
Rad51 activates polyomavirus JC early transcription.PLoS One. 2014 Oct 13;9(10):e110122. doi: 10.1371/journal.pone.0110122. eCollection 2014. PLoS One. 2014. PMID: 25310191 Free PMC article.
-
The Role of the JC Virus in Central Nervous System Tumorigenesis.Int J Mol Sci. 2020 Aug 28;21(17):6236. doi: 10.3390/ijms21176236. Int J Mol Sci. 2020. PMID: 32872288 Free PMC article. Review.
-
Traffic of JC virus from sites of initial infection to the brain: the path to progressive multifocal leukoencephalopathy.J Infect Dis. 2002 Dec 1;186 Suppl 2:S180-6. doi: 10.1086/344280. J Infect Dis. 2002. PMID: 12424695 Review.
Cited by
-
Cell membrane-bound Kaposi's sarcoma-associated herpesvirus-encoded glycoprotein B promotes virus latency by regulating expression of cellular Egr-1.J Biol Chem. 2010 Nov 26;285(48):37491-502. doi: 10.1074/jbc.M110.159103. Epub 2010 Sep 23. J Biol Chem. 2010. PMID: 20864524 Free PMC article.
-
Examining the role of EGR1 during viral infections.Front Microbiol. 2022 Oct 21;13:1020220. doi: 10.3389/fmicb.2022.1020220. eCollection 2022. Front Microbiol. 2022. PMID: 36338037 Free PMC article. Review.
-
JC virus in the pathogenesis of colorectal cancer, an etiological agent or another component in a multistep process?Virol J. 2010 Feb 18;7:42. doi: 10.1186/1743-422X-7-42. Virol J. 2010. PMID: 20167111 Free PMC article. Review.
-
JCPyV NCCR analysis in PML patients with different risk factors: exploring common rearrangements as essential changes for neuropathogenesis.Virol J. 2020 Feb 11;17(1):23. doi: 10.1186/s12985-020-1295-5. Virol J. 2020. PMID: 32046748 Free PMC article.
-
Modulation of JC virus transcription by C/EBPbeta.Virus Res. 2009 Dec;146(1-2):97-106. doi: 10.1016/j.virusres.2009.09.005. Epub 2009 Sep 9. Virus Res. 2009. PMID: 19747512 Free PMC article.
References
-
- Berger JR, Houff S. Progressive multifocal leukoencephalopathy: lessons from AIDS and natalizumab. Neurol Res. 2006;28:299–305. - PubMed
-
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. - PubMed
-
- Chang CF, Otte J, Kerr DA, Valkkila M, Calkins CE, Khalili K. Evidence that the soluble factors secreted by activated immune cells suppress replication of human neurotropic JC virus DNA in glial cells. Virology. 1996;221:226–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

