Allostery: absence of a change in shape does not imply that allostery is not at play
- PMID: 18353365
- PMCID: PMC2684958
- DOI: 10.1016/j.jmb.2008.02.034
Allostery: absence of a change in shape does not imply that allostery is not at play
Abstract
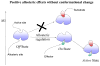
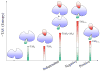
Allostery is essential for controlled catalysis, signal transmission, receptor trafficking, turning genes on and off, and apoptosis. It governs the organism's response to environmental and metabolic cues, dictating transient partner interactions in the cellular network. Textbooks taught us that allostery is a change of shape at one site on the protein surface brought about by ligand binding to another. For several years, it has been broadly accepted that the change of shape is not induced; rather, it is observed simply because a larger protein population presents it. Current data indicate that while side chains can reorient and rewire, allostery may not even involve a change of (backbone) shape. Assuming that the enthalpy change does not reverse the free-energy change due to the change in entropy, entropy is mainly responsible for binding.
Figures

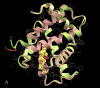
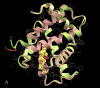
Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Identification of a novel toxicophore in anti-cancer chemotherapeutics that targets mitochondrial respiratory complex I.Elife. 2020 May 20;9:e55845. doi: 10.7554/eLife.55845. Elife. 2020. PMID: 32432547 Free PMC article.
-
Infliximab for maintenance of medically-induced remission in Crohn's disease.Cochrane Database Syst Rev. 2024 Feb 19;2(2):CD012609. doi: 10.1002/14651858.CD012609.pub2. Cochrane Database Syst Rev. 2024. PMID: 38372447 Free PMC article. Review.
-
How to create accessible research summaries for the developmental language disorder community.Int J Lang Commun Disord. 2025 Jan-Feb;60(1):e13142. doi: 10.1111/1460-6984.13142. Int J Lang Commun Disord. 2025. PMID: 39625399
Cited by
-
Probing molecular mechanisms of the Hsp90 chaperone: biophysical modeling identifies key regulators of functional dynamics.PLoS One. 2012;7(5):e37605. doi: 10.1371/journal.pone.0037605. Epub 2012 May 18. PLoS One. 2012. PMID: 22624053 Free PMC article.
-
Engineering allosteric control to an unregulated enzyme by transfer of a regulatory domain.Proc Natl Acad Sci U S A. 2013 Feb 5;110(6):2111-6. doi: 10.1073/pnas.1217923110. Epub 2013 Jan 23. Proc Natl Acad Sci U S A. 2013. PMID: 23345433 Free PMC article.
-
Insights into the activation mechanism of class I HDAC complexes by inositol phosphates.Nat Commun. 2016 Apr 25;7:11262. doi: 10.1038/ncomms11262. Nat Commun. 2016. PMID: 27109927 Free PMC article.
-
Exploration of the conformational landscape in pregnane X receptor reveals a new binding pocket.Protein Sci. 2016 Nov;25(11):1989-2005. doi: 10.1002/pro.3012. Epub 2016 Aug 23. Protein Sci. 2016. PMID: 27515410 Free PMC article.
-
Site-specific 2D IR spectroscopy: a general approach for the characterization of protein dynamics with high spatial and temporal resolution.Phys Chem Chem Phys. 2019 Jan 2;21(2):780-788. doi: 10.1039/c8cp06146g. Phys Chem Chem Phys. 2019. PMID: 30548035 Free PMC article.
References
-
- Daily MD, Gray JJ. Local motions in a benchmark of allosteric proteins. Proteins Struct Funct Genet. 2007;67:385–399. - PubMed
-
- Swain JF, Gierasch LM. The changing landscape of protein allostery. Curr Opin Struct Biol. 2006;16:102–108. - PubMed
-
- Gunasekaran K, Ma BY, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins Struct Funct Genet. 2004;57:433–443. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

