Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter
- PMID: 18321244
- PMCID: PMC2474557
- DOI: 10.1042/BJ20071512
Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6/MKP-3 gene promoter
Abstract
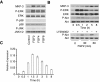
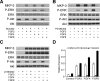
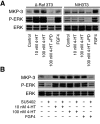
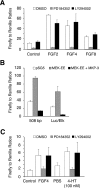
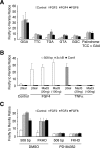
DUSP6 (dual-specificity phosphatase 6), also known as MKP-3 [MAPK (mitogen-activated protein kinase) phosphatase-3] specifically inactivates ERK1/2 (extracellular-signal-regulated kinase 1/2) in vitro and in vivo. DUSP6/MKP-3 is inducible by FGF (fibroblast growth factor) signalling and acts as a negative regulator of ERK activity in key and discrete signalling centres that direct outgrowth and patterning in early vertebrate embryos. However, the molecular mechanism by which FGFs induce DUSP6/MKP-3 expression and hence help to set ERK1/2 signalling levels is unknown. In the present study, we demonstrate, using pharmacological inhibitors and analysis of the murine DUSP6/MKP-3 gene promoter, that the ERK pathway is critical for FGF-induced DUSP6/MKP-3 transcription. Furthermore, we show that this response is mediated by a conserved binding site for the Ets (E twenty-six) family of transcriptional regulators and that the Ets2 protein, a known target of ERK signalling, binds to the endogenous DUSP6/MKP-3 promoter. Finally, the murine DUSP6/MKP-3 promoter coupled to EGFP (enhanced green fluorescent protein) recapitulates the specific pattern of endogenous DUSP6/MKP-3 mRNA expression in the chicken neural plate, where its activity depends on FGFR (FGF receptor) and MAPK signalling and an intact Ets-binding site. These findings identify a conserved Ets-factor-dependent mechanism by which ERK signalling activates DUSP6/MKP-3 transcription to deliver ERK1/2-specific negative-feedback control of FGF signalling.
Figures



Similar articles
-
DUSP6/MKP-3 inactivates ERK1/2 but fails to bind and inactivate ERK5.Cell Signal. 2008 May;20(5):836-43. doi: 10.1016/j.cellsig.2007.12.014. Epub 2007 Dec 27. Cell Signal. 2008. PMID: 18280112
-
Feedback regulation of DUSP6 transcription responding to MAPK1 via ETS2 in human cells.Biochem Biophys Res Commun. 2008 Dec 5;377(1):317-20. doi: 10.1016/j.bbrc.2008.10.003. Epub 2008 Oct 9. Biochem Biophys Res Commun. 2008. PMID: 18848526
-
Fibroblast growth factor receptor signaling activates the human interstitial collagenase promoter via the bipartite Ets-AP1 element.Mol Endocrinol. 1997 Jul;11(8):1129-44. doi: 10.1210/mend.11.8.9958. Mol Endocrinol. 1997. PMID: 9212060
-
Working without kinase activity: phosphotransfer-independent functions of extracellular signal-regulated kinases.Sci Signal. 2011 Oct 25;4(196):re3. doi: 10.1126/scisignal.2002324. Sci Signal. 2011. PMID: 22028468 Review.
-
Dual-specificity MAP kinase phosphatases (MKPs) and cancer.Cancer Metastasis Rev. 2008 Jun;27(2):253-61. doi: 10.1007/s10555-008-9123-1. Cancer Metastasis Rev. 2008. PMID: 18330678 Review.
Cited by
-
The Fibroblast Growth Factor signaling pathway.Wiley Interdiscip Rev Dev Biol. 2015 May-Jun;4(3):215-66. doi: 10.1002/wdev.176. Epub 2015 Mar 13. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25772309 Free PMC article. Review.
-
Regulation of ERK basal and pulsatile activity control proliferation and exit from the stem cell compartment in mammalian epidermis.Proc Natl Acad Sci U S A. 2020 Jul 28;117(30):17796-17807. doi: 10.1073/pnas.2006965117. Epub 2020 Jul 10. Proc Natl Acad Sci U S A. 2020. PMID: 32651268 Free PMC article.
-
Keratinocyte growth factor induces gene expression signature associated with suppression of malignant phenotype of cutaneous squamous carcinoma cells.PLoS One. 2012;7(3):e33041. doi: 10.1371/journal.pone.0033041. Epub 2012 Mar 12. PLoS One. 2012. PMID: 22427941 Free PMC article.
-
WNT/β-catenin modulates the axial identity of embryonic stem cell-derived human neural crest.Development. 2019 Aug 29;146(16):dev175604. doi: 10.1242/dev.175604. Development. 2019. PMID: 31399472 Free PMC article.
-
p53 protein-mediated up-regulation of MAP kinase phosphatase 3 (MKP-3) contributes to the establishment of the cellular senescent phenotype through dephosphorylation of extracellular signal-regulated kinase 1/2 (ERK1/2).J Biol Chem. 2015 Jan 9;290(2):1129-40. doi: 10.1074/jbc.M114.590943. Epub 2014 Nov 20. J Biol Chem. 2015. PMID: 25414256 Free PMC article.
References
-
- Camps M., Nichols A., Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. - PubMed
-
- Dickinson R. J., Keyse S. M. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J. Cell Sci. 2006;119:4607–4615. - PubMed
-
- Muda M., Theodosiou A., Rodrigues N., Boschert U., Camps M., Gillieron C., Davies K., Ashworth A., Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J. Biol. Chem. 1996;271:27205–27208. - PubMed
-
- Nichols A., Camps M., Gillieron C., Chabert C., Brunet A., Wilsbacher J., Cobb M., Pouyssegur J., Shaw J. P., Arkinstall S. Substrate recognition domains within extracellular signal-regulated kinase mediate binding and catalytic activation of mitogen-activated protein kinase phosphatase-3. J. Biol. Chem. 2000;275:24613–24621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

