Newly identified genetic risk variants for celiac disease related to the immune response
- PMID: 18311140
- PMCID: PMC2673512
- DOI: 10.1038/ng.102
Newly identified genetic risk variants for celiac disease related to the immune response
Abstract
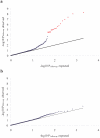
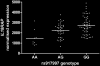
Our genome-wide association study of celiac disease previously identified risk variants in the IL2-IL21 region. To identify additional risk variants, we genotyped 1,020 of the most strongly associated non-HLA markers in an additional 1,643 cases and 3,406 controls. Through joint analysis including the genome-wide association study data (767 cases, 1,422 controls), we identified seven previously unknown risk regions (P < 5 x 10(-7)). Six regions harbor genes controlling immune responses, including CCR3, IL12A, IL18RAP, RGS1, SH2B3 (nsSNP rs3184504) and TAGAP. Whole-blood IL18RAP mRNA expression correlated with IL18RAP genotype. Type 1 diabetes and celiac disease share HLA-DQ, IL2-IL21, CCR3 and SH2B3 risk regions. Thus, this extensive genome-wide association follow-up study has identified additional celiac disease risk variants in relevant biological pathways.
Figures

Similar articles
-
Shared and distinct genetic variants in type 1 diabetes and celiac disease.N Engl J Med. 2008 Dec 25;359(26):2767-77. doi: 10.1056/NEJMoa0807917. Epub 2008 Dec 10. N Engl J Med. 2008. PMID: 19073967 Free PMC article.
-
Six new coeliac disease loci replicated in an Italian population confirm association with coeliac disease.J Med Genet. 2009 Jan;46(1):60-3. doi: 10.1136/jmg.2008.061457. Epub 2008 Sep 19. J Med Genet. 2009. PMID: 18805825
-
Four novel coeliac disease regions replicated in an association study of a Swedish-Norwegian family cohort.Genes Immun. 2010 Jan;11(1):79-86. doi: 10.1038/gene.2009.67. Epub 2009 Aug 20. Genes Immun. 2010. PMID: 19693089
-
Systematic review and meta-analysis of the association between IL18RAP rs917997 and CCR3 rs6441961 polymorphisms with celiac disease risks.Expert Rev Gastroenterol Hepatol. 2015;9(10):1327-38. doi: 10.1586/17474124.2015.1075880. Epub 2015 Aug 8. Expert Rev Gastroenterol Hepatol. 2015. PMID: 26289103 Review.
-
Celiac disease: moving from genetic associations to causal variants.Clin Genet. 2011 Sep;80(3):203-313. doi: 10.1111/j.1399-0004.2011.01707.x. Epub 2011 Jun 13. Clin Genet. 2011. PMID: 21595655 Review.
Cited by
-
High-density genotyping study identifies four new susceptibility loci for atopic dermatitis.Nat Genet. 2013 Jul;45(7):808-12. doi: 10.1038/ng.2642. Epub 2013 Jun 2. Nat Genet. 2013. PMID: 23727859 Free PMC article.
-
Thrombotic antiphospholipid syndrome shows strong haplotypic association with SH2B3-ATXN2 locus.PLoS One. 2013 Jul 3;8(7):e67897. doi: 10.1371/journal.pone.0067897. Print 2013. PLoS One. 2013. PMID: 23844121 Free PMC article.
-
Mining disease genes using integrated protein-protein interaction and gene-gene co-regulation information.FEBS Open Bio. 2015 Mar 27;5:251-6. doi: 10.1016/j.fob.2015.03.011. eCollection 2015. FEBS Open Bio. 2015. PMID: 25870785 Free PMC article.
-
Genomic prediction of celiac disease targeting HLA-positive individuals.Genome Med. 2015 Jul 16;7(1):72. doi: 10.1186/s13073-015-0196-5. eCollection 2015. Genome Med. 2015. PMID: 26244058 Free PMC article.
-
Data-driven modelling of mutational hotspots and in silico predictors in hypertrophic cardiomyopathy.J Med Genet. 2021 Aug;58(8):556-564. doi: 10.1136/jmedgenet-2020-106922. Epub 2020 Jul 30. J Med Genet. 2021. PMID: 32732227 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

