Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder
- PMID: 18299402
- PMCID: PMC2275382
- DOI: 10.1084/jem.20072108
Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder
Abstract
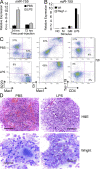
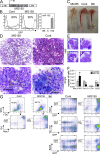
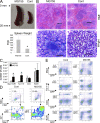
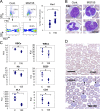
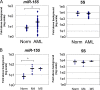
Mammalian microRNAs are emerging as key regulators of the development and function of the immune system. Here, we report a strong but transient induction of miR-155 in mouse bone marrow after injection of bacterial lipopolysaccharide (LPS) correlated with granulocyte/monocyte (GM) expansion. Demonstrating the sufficiency of miR-155 to drive GM expansion, enforced expression in mouse bone marrow cells caused GM proliferation in a manner reminiscent of LPS treatment. However, the miR-155-induced GM populations displayed pathological features characteristic of myeloid neoplasia. Of possible relevance to human disease, miR-155 was found to be overexpressed in the bone marrow of patients with certain subtypes of acute myeloid leukemia (AML). Furthermore, miR-155 repressed a subset of genes implicated in hematopoietic development and disease. These data implicate miR-155 as a contributor to physiological GM expansion during inflammation and to certain pathological features associated with AML, emphasizing the importance of proper miR-155 regulation in developing myeloid cells during times of inflammatory stress.
Figures


Similar articles
-
Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis.Blood. 2009 Apr 30;113(18):4414-24. doi: 10.1182/blood-2008-10-182626. Epub 2009 Jan 29. Blood. 2009. PMID: 19179468 Free PMC article.
-
MicroRNA-143 targets ERK5 in granulopoiesis and predicts outcome of patients with acute myeloid leukemia.Cell Death Dis. 2018 Jul 26;9(8):814. doi: 10.1038/s41419-018-0837-x. Cell Death Dis. 2018. PMID: 30050105 Free PMC article.
-
MicroRNA-9 promotes proliferation of leukemia cells in adult CD34-positive acute myeloid leukemia with normal karyotype by downregulation of Hes1.Tumour Biol. 2016 Jun;37(6):7461-71. doi: 10.1007/s13277-015-4581-x. Epub 2015 Dec 17. Tumour Biol. 2016. PMID: 26678889
-
MicroRNAs expressed in hematopoietic stem/progenitor cells are deregulated in acute myeloid leukemias.Leuk Lymphoma. 2015 May;56(5):1466-74. doi: 10.3109/10428194.2014.955019. Epub 2015 Jan 21. Leuk Lymphoma. 2015. PMID: 25242094 Review.
-
The role of HMGA2 in the proliferation and expansion of a hematopoietic cell in myeloproliferative neoplasms.Fukushima J Med Sci. 2012;58(2):91-100. doi: 10.5387/fms.58.91. Fukushima J Med Sci. 2012. PMID: 23237864 Review.
Cited by
-
MicroRNAs as mediators of viral evasion of the immune system.Nat Immunol. 2013 Mar;14(3):205-10. doi: 10.1038/ni.2537. Epub 2013 Feb 15. Nat Immunol. 2013. PMID: 23416678 Free PMC article. Review.
-
Macrophage polarization and plasticity in health and disease.Immunol Res. 2012 Sep;53(1-3):11-24. doi: 10.1007/s12026-012-8291-9. Immunol Res. 2012. PMID: 22418728 Review.
-
Mir-55 inhibition can reduce cell proliferation and induce apoptosis in Jurkat (Acute T cell Leukemia) cell line.Iran J Ped Hematol Oncol. 2014;4(4):141-50. Epub 2014 Dec 10. Iran J Ped Hematol Oncol. 2014. PMID: 25598954 Free PMC article.
-
MicroRNAs in human cancer.Adv Exp Med Biol. 2013;774:1-20. doi: 10.1007/978-94-007-5590-1_1. Adv Exp Med Biol. 2013. PMID: 23377965 Free PMC article. Review.
-
Molecular signatures of maturing dendritic cells: implications for testing the quality of dendritic cell therapies.J Transl Med. 2010 Jan 15;8:4. doi: 10.1186/1479-5876-8-4. J Transl Med. 2010. PMID: 20078880 Free PMC article.
References
-
- Rosenbauer, F., and D.G. Tenen. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117. - PubMed
-
- Ambros, V. 2004. The functions of animal microRNAs. Nature. 431:350–355. - PubMed
-
- Bartel, D.P., and C.Z. Chen. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5:396–400. - PubMed
-
- He, L., and G.J. Hannon. 2004. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5:522–531. - PubMed
-
- Georgantas, R.W. III, R. Hildreth, S. Morisot, J. Alder, C.G. Liu, S. Heimfeld, G.A. Calin, C.M. Croce, and C.I. Civin. 2007. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA. 104:2750–2755. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

