Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria
- PMID: 18296640
- PMCID: PMC2268797
- DOI: 10.1073/pnas.0712209105
Selective association of misfolded ALS-linked mutant SOD1 with the cytoplasmic face of mitochondria
Abstract
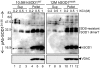
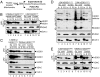
Mutations in copper/zinc superoxide dismutase (SOD1) are causative for dominantly inherited amyotrophic lateral sclerosis (ALS). Despite high variability in biochemical properties among the disease-causing mutants, a proportion of both dismutase-active and -inactive mutants are stably bound to spinal cord mitochondria. This mitochondrial proportion floats with mitochondria rather than sedimenting to the much higher density of protein, thus eliminating coincidental cosedimentation of protein aggregates with mitochondria. Half of dismutase-active and approximately 90% of dismutase-inactive mutant SOD1 is bound to mitochondrial membranes in an alkali- and salt-resistant manner. Sensitivity to proteolysis and immunoprecipitation with an antibody specific for misfolded SOD1 demonstrate that in all mutant SOD1 models, misfolded SOD1 is deposited onto the cytoplasmic face of the outer mitochondrial membrane, increasing antigenic accessibility of the normally structured electrostatic loop. Misfolded mutant SOD1 binding is both restricted to spinal cord and selective for mitochondrial membranes, implicating exposure to mitochondria of a misfolded mutant SOD1 conformer mediated by a unique, tissue-selective composition of cytoplasmic chaperones, components unique to the cytoplasmic face of spinal mitochondria to which misfolded SOD1 binds, or misfolded SOD1 conformers unique to spinal cord that have a selective affinity for mitochondrial membranes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

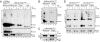

Similar articles
-
Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space.Antioxid Redox Signal. 2010 Nov 1;13(9):1375-84. doi: 10.1089/ars.2010.3212. Antioxid Redox Signal. 2010. PMID: 20367259 Free PMC article. Review.
-
Proteins that bind to misfolded mutant superoxide dismutase-1 in spinal cords from transgenic amyotrophic lateral sclerosis (ALS) model mice.J Biol Chem. 2011 Jun 10;286(23):20130-6. doi: 10.1074/jbc.M111.218842. Epub 2011 Apr 14. J Biol Chem. 2011. PMID: 21493711 Free PMC article.
-
Mutant SOD1 from spinal cord of G93A rats is destabilized and binds to inner mitochondrial membrane.Neurobiol Dis. 2008 Dec;32(3):479-85. doi: 10.1016/j.nbd.2008.08.010. Epub 2008 Sep 9. Neurobiol Dis. 2008. PMID: 18817872
-
Toxicity of familial ALS-linked SOD1 mutants from selective recruitment to spinal mitochondria.Neuron. 2004 Jul 8;43(1):5-17. doi: 10.1016/j.neuron.2004.06.016. Neuron. 2004. PMID: 15233913
-
Oxidized/misfolded superoxide dismutase-1: the cause of all amyotrophic lateral sclerosis?Ann Neurol. 2007 Dec;62(6):553-9. doi: 10.1002/ana.21319. Ann Neurol. 2007. PMID: 18074357 Review.
Cited by
-
ASCs-Exosomes Recover Coupling Efficiency and Mitochondrial Membrane Potential in an in vitro Model of ALS.Front Neurosci. 2019 Oct 17;13:1070. doi: 10.3389/fnins.2019.01070. eCollection 2019. Front Neurosci. 2019. PMID: 31680811 Free PMC article.
-
Import, maturation, and function of SOD1 and its copper chaperone CCS in the mitochondrial intermembrane space.Antioxid Redox Signal. 2010 Nov 1;13(9):1375-84. doi: 10.1089/ars.2010.3212. Antioxid Redox Signal. 2010. PMID: 20367259 Free PMC article. Review.
-
Dysregulation of the proteasome increases the toxicity of ALS-linked mutant SOD1.Genes Cells. 2014 Mar;19(3):209-24. doi: 10.1111/gtc.12125. Epub 2014 Jan 23. Genes Cells. 2014. PMID: 24450587 Free PMC article.
-
The role of environmental exposures in neurodegeneration and neurodegenerative diseases.Toxicol Sci. 2011 Dec;124(2):225-50. doi: 10.1093/toxsci/kfr239. Epub 2011 Sep 13. Toxicol Sci. 2011. PMID: 21914720 Free PMC article. Review.
-
Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis.Nat Rev Neurol. 2011 Nov;7(11):616-30. doi: 10.1038/nrneurol.2011.152. Nat Rev Neurol. 2011. PMID: 22051914 Review.
References
-
- Boillee S, Vande Velde C, Cleveland DW. ALS: A disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:1–21. - PubMed
-
- Bruijn LI, et al. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. - PubMed
-
- Bruijn LI, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. - PubMed
-
- Reaume AG, et al. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat Genet. 1996;13:43–47. - PubMed
-
- Hirano A, Donnenfeld H, Sasaki S, Nakano I. Fine structural observations of neurofilamentous changes in amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 1984;43:461–470. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

