Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles
- PMID: 18295294
- PMCID: PMC2443636
- DOI: 10.1016/j.virol.2008.01.018
Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles
Abstract
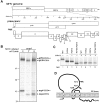
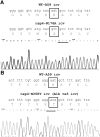
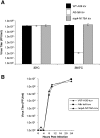
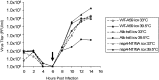
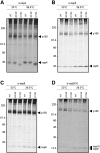
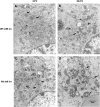
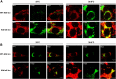
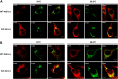
Coronaviruses are positive-strand RNA viruses that replicate in the cytoplasm of infected cells by generating a membrane-associated replicase complex. The replicase complex assembles on double membrane vesicles (DMVs). Here, we studied the role of a putative replicase anchor, nonstructural protein 4 (nsp4), in the assembly of murine coronavirus DMVs. We used reverse genetics to generate infectious clone viruses (icv) with an alanine substitution at nsp4 glycosylation site N176 or N237, or an asparagine to threonine substitution (nsp4-N258T), which is proposed to confer a temperature sensitive phenotype. We found that nsp4-N237A is lethal and nsp4-N258T generated a virus (designated Alb ts6 icv) that is temperature sensitive for viral replication. Analysis of Alb ts6 icv-infected cells revealed that there was a dramatic reduction in DMVs and that both nsp4 and nsp3 partially localized to mitochondria when cells were incubated at the non-permissive temperature. These results reveal a critical role of nsp4 in directing coronavirus DMV assembly.
Figures
Similar articles
-
Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function.J Virol. 2010 Jan;84(1):280-90. doi: 10.1128/JVI.01772-09. J Virol. 2010. PMID: 19846526 Free PMC article.
-
Mutations across murine hepatitis virus nsp4 alter virus fitness and membrane modifications.J Virol. 2015 Feb;89(4):2080-9. doi: 10.1128/JVI.02776-14. Epub 2014 Dec 3. J Virol. 2015. PMID: 25473044 Free PMC article.
-
Expression and Cleavage of Middle East Respiratory Syndrome Coronavirus nsp3-4 Polyprotein Induce the Formation of Double-Membrane Vesicles That Mimic Those Associated with Coronaviral RNA Replication.mBio. 2017 Nov 21;8(6):e01658-17. doi: 10.1128/mBio.01658-17. mBio. 2017. PMID: 29162711 Free PMC article.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
Rotavirus protein expression is important for virus assembly and pathogenesis.Arch Virol Suppl. 1996;12:69-77. doi: 10.1007/978-3-7091-6553-9_8. Arch Virol Suppl. 1996. PMID: 9015103 Review.
Cited by
-
Assay Development and Validation for Innovative Antiviral Development Targeting the N-Terminal Autoprocessing of SARS-CoV-2 Main Protease Precursors.Viruses. 2024 Jul 29;16(8):1218. doi: 10.3390/v16081218. Viruses. 2024. PMID: 39205192 Free PMC article.
-
Evidence of mitochondria origin of SARS-CoV-2 double-membrane vesicles: a review.F1000Res. 2024 Jul 10;10:1009. doi: 10.12688/f1000research.73170.2. eCollection 2021. F1000Res. 2024. PMID: 38827572 Free PMC article. Review.
-
Expression and immunogenicity of non-structural protein 8 of porcine epidemic diarrhea virus.Vet Res Forum. 2024;15(2):65-73. doi: 10.30466/vrf.2023.2009322.3977. Epub 2024 Feb 15. Vet Res Forum. 2024. PMID: 38465319 Free PMC article.
-
Nsp3-N interactions are critical for SARS-CoV-2 fitness and virulence.Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2305674120. doi: 10.1073/pnas.2305674120. Epub 2023 Jul 24. Proc Natl Acad Sci U S A. 2023. PMID: 37487098 Free PMC article.
-
Endomembrane remodeling in SARS-CoV-2 infection.Cell Insight. 2022 May 17;1(3):100031. doi: 10.1016/j.cellin.2022.100031. eCollection 2022 Jun. Cell Insight. 2022. PMID: 37193051 Free PMC article. Review.
References
-
- Baker S.C., Denison M.R. Cell biology of nidovirus replication complexes. In: Perlman S., Gallagher T., Snijder E., editors. Nidoviruses. ASM Press; Washington: 2008. pp. 103–113.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

