Epigenetic inactivation of RASSF2 in oral squamous cell carcinoma
- PMID: 18294275
- PMCID: PMC11158976
- DOI: 10.1111/j.1349-7006.2008.00769.x
Epigenetic inactivation of RASSF2 in oral squamous cell carcinoma
Abstract
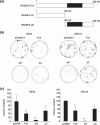
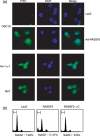
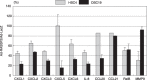
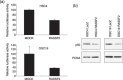
Genetic and epigenetic alterations in tumor-suppressor genes play important roles in human neoplasia. Ras signaling is often activated in oral squamous cell carcinoma (OSCC), although Ras mutations are rarely detected in Japanese OSCC patients, and the mechanisms underlying the gene's activation remain unclear. Here, we examined the expression of Ras association family (RASSF) genes in a panel of OSCC cell lines and found that RASSF2 is often downregulated by DNA methylation in OSCC cells. In addition, aberrant methylation of RASSF2 was detected in 12 of 46 (26%) primary OSCC, and 18 (39%) of those OSCC showed methylation of at least one RASSF gene. Ectopic expression of RASSF2 in OSCC cells suppressed cell growth and induced apoptosis. A RASSF2 deletion mutant lacking the Ras-association domain, which was therefore unable to interact with Ras, exhibited less pro-apoptotic activity than the full-length protein, indicating that the pro-apoptotic activity of RASSF2 is related to its association with Ras. Genomic screening of genes regulated by RASSF2 showed that genes involved in immune responses, angiogenesis, and metastasis are suppressed by RASSF2. Our results suggest that epigenetic inactivation of RASSF2 plays an important role in OSCC tumorigenesis, and that RASSF2 may be a useful molecular target for the diagnosis and treatment of OSCC.
Figures



Similar articles
-
The Ras effector RASSF2 is a novel tumor-suppressor gene in human colorectal cancer.Gastroenterology. 2005 Jul;129(1):156-69. doi: 10.1053/j.gastro.2005.03.051. Gastroenterology. 2005. PMID: 16012945
-
Epigenetic inactivation of the RAS-effector gene RASSF2 in lung cancers.Int J Oncol. 2007 Jul;31(1):169-73. Int J Oncol. 2007. PMID: 17549418
-
Epigenetic inactivation of SFRP genes in oral squamous cell carcinoma.Int J Oncol. 2008 Jun;32(6):1253-61. doi: 10.3892/ijo_32_6_1253. Int J Oncol. 2008. PMID: 18497987
-
DNA methylation in oral squamous cell carcinoma: molecular mechanisms and clinical implications.Oral Dis. 2011 Nov;17(8):771-8. doi: 10.1111/j.1601-0825.2011.01833.x. Epub 2011 Jul 22. Oral Dis. 2011. PMID: 21781230 Review.
-
Epigenetic mechanisms in oral carcinogenesis.Future Oncol. 2012 Nov;8(11):1407-25. doi: 10.2217/fon.12.138. Future Oncol. 2012. PMID: 23148615 Free PMC article. Review.
Cited by
-
Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review.Cancers (Basel). 2023 Nov 27;15(23):5600. doi: 10.3390/cancers15235600. Cancers (Basel). 2023. PMID: 38067304 Free PMC article. Review.
-
Anti-Proliferative Effect of Statins Is Mediated by DNMT1 Inhibition and p21 Expression in OSCC Cells.Cancers (Basel). 2020 Jul 28;12(8):2084. doi: 10.3390/cancers12082084. Cancers (Basel). 2020. PMID: 32731382 Free PMC article.
-
Phospholipase C epsilon plays a suppressive role in incidence of colorectal cancer.Med Oncol. 2012 Jun;29(2):1051-8. doi: 10.1007/s12032-011-9981-1. Epub 2011 Jun 11. Med Oncol. 2012. PMID: 21667163
-
Ablation of Rassf2 induces bone defects and subsequent haematopoietic anomalies in mice.EMBO J. 2012 Mar 7;31(5):1147-59. doi: 10.1038/emboj.2011.480. Epub 2012 Jan 6. EMBO J. 2012. PMID: 22227519 Free PMC article.
-
Ras signaling through RASSF proteins.Semin Cell Dev Biol. 2016 Oct;58:86-95. doi: 10.1016/j.semcdb.2016.06.007. Epub 2016 Jun 8. Semin Cell Dev Biol. 2016. PMID: 27288568 Free PMC article. Review.
References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108. - PubMed
-
- Gangane N, Chawla S, Subodh A, Gupta S, Sharma SM. Reassessment of risk factors for oral cancer. Asian Pac J Cancer Prev 2007; 8: 243–8. - PubMed
-
- Petti S, Scully C. Oral cancer: the association between nation‐based alcohol‐drinking profiles and oral cancer mortality. Oral Oncol 2005; 41: 828–34. - PubMed
-
- Chiba I, Shindoh M, Yasuda M et al . Mutations in the p53 gene and human papillomavirus infection as significant prognostic factors in squamous cell carcinomas of the oral cavity. Oncogene 1996; 12: 1663–8. - PubMed
-
- Ha PK, Califano JA. Promoter methylation and inactivation of tumour‐suppressor genes in oral squamous‐cell carcinoma. Lancet Oncol 2006; 7: 77–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

