G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II
- PMID: 18292094
- PMCID: PMC2442332
- DOI: 10.1074/jbc.M705003200
G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II
Abstract
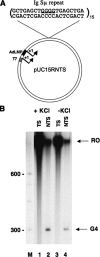
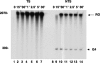
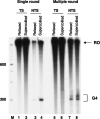
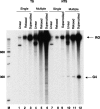
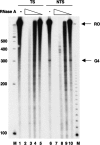
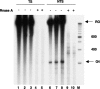
DNA sequences rich in runs of guanine have the potential to form G4 DNA, a four-stranded non-canonical DNA structure stabilized by formation and stacking of G quartets, planar arrays of four hydrogen-bonded guanines. It was reported recently that G4 DNA can be generated in Escherichia coli during transcription of plasmids containing G-rich sequences in the non-transcribed strand. In addition, a stable RNA/DNA hybrid is formed with the transcribed strand. These novel structures, termed G loops, are suppressed in recQ(+) strains, suggesting that their persistence may generate genomic instability and that the RecQ helicase may be involved in their dissolution. However, little is known about how such non-canonical DNA structures are processed when encountered by an elongating polymerase. To assess whether G4-forming sequences interfere with transcription, we studied their effect on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. We used a reconstituted transcription system in vitro with purified polymerase and initiation factors and with substrates containing G-rich sequences in either the transcribed or non-transcribed strand downstream of the T7 promoter or the adenovirus major late promoter. We report that G-rich sequences located in the transcribed strand do not affect transcription by either polymerase, but when the sequences are located in the non-transcribed strand, they partially arrest both polymerases. The efficiency of arrest increases with negative supercoiling and also with multiple rounds of transcription compared with single events.
Figures

Similar articles
-
Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II.DNA Repair (Amst). 2004 May 4;3(5):483-94. doi: 10.1016/j.dnarep.2004.01.003. DNA Repair (Amst). 2004. PMID: 15084310
-
Transcription arrest at a lesion in the transcribed DNA strand in vitro is not affected by a nearby lesion in the opposite strand.J Biol Chem. 2003 May 23;278(21):19558-64. doi: 10.1074/jbc.M301060200. Epub 2003 Mar 19. J Biol Chem. 2003. PMID: 12646562
-
Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II.J Biol Chem. 2001 Nov 30;276(48):45367-71. doi: 10.1074/jbc.M105282200. Epub 2001 Sep 24. J Biol Chem. 2001. PMID: 11571287 Free PMC article.
-
[Promoters of eukaryotic genes transcribed by RNA polymerase II].Biochimie. 1982 Oct;64(10):III-VII. Biochimie. 1982. PMID: 6817816 Review. French. No abstract available.
-
RNA polymerase collisions and their role in transcription.Transcription. 2024 Feb-Apr;15(1-2):38-47. doi: 10.1080/21541264.2024.2316972. Epub 2024 Feb 15. Transcription. 2024. PMID: 38357902 Review.
Cited by
-
An Upstream G-Quadruplex DNA Structure Can Stimulate Gene Transcription.ACS Chem Biol. 2024 Mar 15;19(3):736-742. doi: 10.1021/acschembio.3c00775. Epub 2024 Feb 28. ACS Chem Biol. 2024. PMID: 38417105 Free PMC article.
-
RNA polymerase pausing, stalling and bypass during transcription of damaged DNA: from molecular basis to functional consequences.Nucleic Acids Res. 2022 Apr 8;50(6):3018-3041. doi: 10.1093/nar/gkac174. Nucleic Acids Res. 2022. PMID: 35323981 Free PMC article. Review.
-
Molecular basis of transcriptional pausing, stalling, and transcription-coupled repair initiation.Biochim Biophys Acta Gene Regul Mech. 2021 Jan;1864(1):194659. doi: 10.1016/j.bbagrm.2020.194659. Epub 2020 Nov 30. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 33271312 Free PMC article. Review.
-
The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease.Nat Rev Neurosci. 2016 Jun;17(6):383-95. doi: 10.1038/nrn.2016.38. Epub 2016 May 6. Nat Rev Neurosci. 2016. PMID: 27150398 Free PMC article. Review.
-
DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication.Biochim Biophys Acta. 2013 Aug;1829(8):854-65. doi: 10.1016/j.bbagrm.2013.03.012. Epub 2013 Apr 6. Biochim Biophys Acta. 2013. PMID: 23567047 Free PMC article. Review.
References
-
- Sinden, R. R. (1994) DNA Structure and Function, Academic Press, San Diego, CA
-
- Sen, D., and Gilbert, W. (1988) Nature 334 364–366 - PubMed
-
- Duquette, M. L., Pham, P., Goodman, M. F., and Maizels, N. (2005) Oncogene 24 5791–5798 - PubMed
-
- Paeschke, K., Simonsson, T., Postberg, J., Rhodes, D., and Lipps, H. J. (2005) Nat. Struct. Mol. Biol. 12 847–854 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

