A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response
- PMID: 18287073
- PMCID: PMC2268563
- DOI: 10.1073/pnas.0708670105
A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response
Abstract
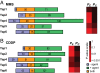
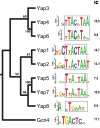
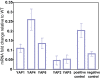
Duplication of genes encoding transcription factors plays an essential role in driving phenotypic variation. Because regulation can occur at multiple levels, it is often difficult to discern how each duplicated factor achieves its regulatory specificity. In these cases, a "systems approach" may distinguish the role of each factor by integrating complementary large-scale measurements of the regulatory network. To explore such an approach, we integrate growth phenotypes, promoter binding profiles, and gene expression patterns to model the DNA damage response network controlled by the Yeast-specific AP-1 (YAP) family of transcription factors. This analysis reveals that YAP regulatory specificity is achieved by at least three mechanisms: (i) divergence of DNA-binding sequences into two subfamilies; (ii) condition-specific combinatorial regulation by multiple Yap factors; and (iii) interactions of Yap 1, 4, and 6 with chromatin remodeling proteins. Additional microarray experiments establish that Yap 4 and 6 regulate gene expression through interactions with the histone deacetylase, Hda1. The data further highlight differences among Yap paralogs in terms of their regulatory mode of action (activation vs. repression). This study suggests how other large TF families might be disentangled in the future.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Genome-Wide Mapping Targets of the Metazoan Chromatin Remodeling Factor NURF Reveals Nucleosome Remodeling at Enhancers, Core Promoters and Gene Insulators.PLoS Genet. 2016 Apr 5;12(4):e1005969. doi: 10.1371/journal.pgen.1005969. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27046080 Free PMC article.
-
Runx2/Cbfa1 functions: diverse regulation of gene transcription by chromatin remodeling and co-regulatory protein interactions.Connect Tissue Res. 2003;44 Suppl 1:141-8. Connect Tissue Res. 2003. PMID: 12952188 Review.
-
YAP/TAZ regulates TGF-β/Smad3 signaling by induction of Smad7 via AP-1 in human skin dermal fibroblasts.Cell Commun Signal. 2018 Apr 25;16(1):18. doi: 10.1186/s12964-018-0232-3. Cell Commun Signal. 2018. PMID: 29695252 Free PMC article.
-
Human INO80 chromatin-remodelling complex contributes to DNA double-strand break repair via the expression of Rad54B and XRCC3 genes.Biochem J. 2010 Oct 15;431(2):179-87. doi: 10.1042/BJ20100988. Biochem J. 2010. PMID: 20687897
-
The transcriptional regulatory code of eukaryotic cells--insights from genome-wide analysis of chromatin organization and transcription factor binding.Curr Opin Cell Biol. 2006 Jun;18(3):291-8. doi: 10.1016/j.ceb.2006.04.002. Epub 2006 May 2. Curr Opin Cell Biol. 2006. PMID: 16647254 Review.
Cited by
-
An atlas of combinatorial transcriptional regulation in mouse and man.Cell. 2010 Mar 5;140(5):744-52. doi: 10.1016/j.cell.2010.01.044. Cell. 2010. PMID: 20211142 Free PMC article.
-
Coevolution within a transcriptional network by compensatory trans and cis mutations.Genome Res. 2010 Dec;20(12):1672-8. doi: 10.1101/gr.111765.110. Epub 2010 Oct 26. Genome Res. 2010. PMID: 20978140 Free PMC article.
-
Environment-responsive transcription factors bind subtelomeric elements and regulate gene silencing.Mol Syst Biol. 2011 Jan 4;7:455. doi: 10.1038/msb.2010.110. Mol Syst Biol. 2011. PMID: 21206489 Free PMC article.
-
A Network of Paralogous Stress Response Transcription Factors in the Human Pathogen Candida glabrata.Front Microbiol. 2016 May 9;7:645. doi: 10.3389/fmicb.2016.00645. eCollection 2016. Front Microbiol. 2016. PMID: 27242683 Free PMC article.
-
Determining PTEN functional status by network component deduced transcription factor activities.PLoS One. 2012;7(2):e31053. doi: 10.1371/journal.pone.0031053. Epub 2012 Feb 8. PLoS One. 2012. PMID: 22347425 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

