Analyzing M-CSF dependent monocyte/macrophage differentiation: expression modes and meta-modes derived from an independent component analysis
- PMID: 18279525
- PMCID: PMC2277398
- DOI: 10.1186/1471-2105-9-100
Analyzing M-CSF dependent monocyte/macrophage differentiation: expression modes and meta-modes derived from an independent component analysis
Abstract
Background: The analysis of high-throughput gene expression data sets derived from microarray experiments still is a field of extensive investigation. Although new approaches and algorithms are published continuously, mostly conventional methods like hierarchical clustering algorithms or variance analysis tools are used. Here we take a closer look at independent component analysis (ICA) which is already discussed widely as a new analysis approach. However, deep exploration of its applicability and relevance to concrete biological problems is still missing. In this study, we investigate the relevance of ICA in gaining new insights into well characterized regulatory mechanisms of M-CSF dependent macrophage differentiation.
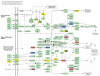
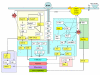
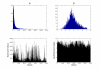
Results: Statistically independent gene expression modes (GEM) were extracted from observed gene expression signatures (GES) through ICA of different microarray experiments. From each GEM we deduced a group of genes, henceforth called sub-mode. These sub-modes were further analyzed with different database query and literature mining tools and then combined to form so called meta-modes. With them we performed a knowledge-based pathway analysis and reconstructed a well known signal cascade.
Conclusion: We show that ICA is an appropriate tool to uncover underlying biological mechanisms from microarray data. Most of the well known pathways of M-CSF dependent monocyte to macrophage differentiation can be identified by this unsupervised microarray data analysis. Moreover, recent research results like the involvement of proliferation associated cellular mechanisms during macrophage differentiation can be corroborated.
Figures


Similar articles
-
How to extract marker genes from microarray data sets.Annu Int Conf IEEE Eng Med Biol Soc. 2007;2007:4215-8. doi: 10.1109/IEMBS.2007.4353266. Annu Int Conf IEEE Eng Med Biol Soc. 2007. PMID: 18002932
-
Analyzing time-dependent microarray data using independent component analysis derived expression modes from human macrophages infected with F. tularensis holartica.J Biomed Inform. 2009 Aug;42(4):605-11. doi: 10.1016/j.jbi.2009.01.002. Epub 2009 Jan 23. J Biomed Inform. 2009. PMID: 19535009
-
An mRNA atlas of G protein-coupled receptor expression during primary human monocyte/macrophage differentiation and lipopolysaccharide-mediated activation identifies targetable candidate regulators of inflammation.Immunobiology. 2013 Nov;218(11):1345-53. doi: 10.1016/j.imbio.2013.07.001. Epub 2013 Jul 15. Immunobiology. 2013. PMID: 23948647
-
The roles of CSFs on the functional polarization of tumor-associated macrophages.FEBS J. 2018 Feb;285(4):680-699. doi: 10.1111/febs.14343. Epub 2017 Dec 16. FEBS J. 2018. PMID: 29171156 Review.
-
CSF-1 signaling in macrophages: pleiotrophy through phosphotyrosine-based signaling pathways.Crit Rev Clin Lab Sci. 2012 Mar-Apr;49(2):49-61. doi: 10.3109/10408363.2012.666845. Crit Rev Clin Lab Sci. 2012. PMID: 22468857 Review.
Cited by
-
K1 and K15 of Kaposi's Sarcoma-Associated Herpesvirus Are Partial Functional Homologues of Latent Membrane Protein 2A of Epstein-Barr Virus.J Virol. 2015 Jul;89(14):7248-61. doi: 10.1128/JVI.00839-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948739 Free PMC article.
-
Is There a Need for a More Precise Description of Biomolecule Interactions to Understand Cell Function?Curr Issues Mol Biol. 2022 Jan 21;44(2):505-525. doi: 10.3390/cimb44020035. Curr Issues Mol Biol. 2022. PMID: 35723321 Free PMC article. Review.
-
Linking Activation of Microglia and Peripheral Monocytic Cells to the Pathophysiology of Psychiatric Disorders.Front Cell Neurosci. 2016 Jun 3;10:144. doi: 10.3389/fncel.2016.00144. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27375431 Free PMC article. Review.
-
Independent component and pathway-based analysis of miRNA-regulated gene expression in a model of type 1 diabetes.BMC Genomics. 2011 Feb 4;12:97. doi: 10.1186/1471-2164-12-97. BMC Genomics. 2011. PMID: 21294859 Free PMC article.
-
Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans.PLoS Genet. 2011 Dec;7(12):e1002367. doi: 10.1371/journal.pgen.1002367. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144904 Free PMC article.
References
-
- Makeigh S, Bell A, Jung TP, Sejnowski T. Advances in Neural Information Processing Systems 8 (NIPS'1995) Cambridge, Masachusetts: MIT Press; 1995. Independent component analysis of electroencephalographic data.
-
- Habl M, Bauer C, Ziegaus C, Lang EW. Perspectives in Neuroscience: Artificial Neural Networks in Medicine and Biology. Springer Publishers Berlin 2000 chap. Analyzing Brain Tumor Related EEG Signals With ICA Algorithms; pp. 131–136.
-
- Vigario R, Jousmäki V, Hämäläinen M, Hari R, Oja E. Advances in Neural Information Processing Systems 10 (NIPS'1997) Cambridge, Masachusetts: MIT Press; 1997. Independent component analysis for identification of artifacts in Magnetoencephalographic recordings; pp. 229–235.
-
- Yang K, Rajapakse JC. ICA gives higher-order functional connectivity of brain. Neural Information Processing – Letters and Reviews. 2004;2:27–32.
-
- Keck IR, Theis FJ, Gruber P, Lang EW, Specht K, Puntonet CG. 3D spatial analysis of fMRI Data on a word perception task. In: Puntonet CG, Prieto A, editor. Lecture Notes in Computer Science, LNCS 3195. Berlin: Springer Verlag; 2004. pp. 977–984.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

