Evolutionary relationships between bat coronaviruses and their hosts
- PMID: 18258002
- PMCID: PMC2851503
- DOI: 10.3201/eid1310.070448
Evolutionary relationships between bat coronaviruses and their hosts
Abstract
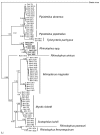
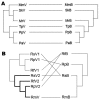
Recent studies have suggested that bats are the natural reservoir of a range of coronaviruses (CoVs), and that rhinolophid bats harbor viruses closely related to the severe acute respiratory syndrome (SARS) CoV, which caused an outbreak of respiratory illness in humans during 2002-2003. We examined the evolutionary relationships between bat CoVs and their hosts by using sequence data of the virus RNA-dependent RNA polymerase gene and the bat cytochrome b gene. Phylogenetic analyses showed multiple incongruent associations between the phylogenies of rhinolophid bats and their CoVs, which suggested that host shifts have occurred in the recent evolutionary history of this group. These shifts may be due to either virus biologic traits or host behavioral traits. This finding has implications for the emergence of SARS and for the potential future emergence of SARS-CoVs or related viruses.
Figures

Similar articles
-
Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History.J Virol. 2017 Feb 14;91(5):e01953-16. doi: 10.1128/JVI.01953-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28077633 Free PMC article.
-
SARS-Coronavirus ancestor's foot-prints in South-East Asian bat colonies and the refuge theory.Infect Genet Evol. 2011 Oct;11(7):1690-702. doi: 10.1016/j.meegid.2011.06.021. Epub 2011 Jul 8. Infect Genet Evol. 2011. PMID: 21763784 Free PMC article.
-
Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China.Emerg Microbes Infect. 2017 Mar 29;6(3):e14. doi: 10.1038/emi.2016.140. Emerg Microbes Infect. 2017. PMID: 28352124 Free PMC article.
-
Geographical structure of bat SARS-related coronaviruses.Infect Genet Evol. 2019 Apr;69:224-229. doi: 10.1016/j.meegid.2019.02.001. Epub 2019 Feb 6. Infect Genet Evol. 2019. PMID: 30735813 Free PMC article. Review.
-
Bats and Coronaviruses.Viruses. 2019 Jan 9;11(1):41. doi: 10.3390/v11010041. Viruses. 2019. PMID: 30634396 Free PMC article. Review.
Cited by
-
New record of Miniopterusmagnater (Chiroptera, Miniopteridae) from south-western China and a comparative study of three species of Miniopterus in China.Biodivers Data J. 2024 Sep 13;12:e129879. doi: 10.3897/BDJ.12.e129879. eCollection 2024. Biodivers Data J. 2024. PMID: 39309533 Free PMC article.
-
Surveillance strategies for SARS-CoV-2 infections through one health approach.Heliyon. 2024 Aug 30;10(17):e37128. doi: 10.1016/j.heliyon.2024.e37128. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39286214 Free PMC article. Review.
-
Host-Virus Cophylogenetic Trajectories: Investigating Molecular Relationships between Coronaviruses and Bat Hosts.Viruses. 2024 Jul 15;16(7):1133. doi: 10.3390/v16071133. Viruses. 2024. PMID: 39066295 Free PMC article.
-
Comparative genomics of spike, envelope, and nucleocapsid protein of severe acute respiratory syndrome coronavirus 2.Afr Health Sci. 2023 Sep;23(3):384-399. doi: 10.4314/ahs.v23i3.45. Afr Health Sci. 2023. PMID: 38357143 Free PMC article.
-
Karyotypic stasis and swarming influenced the evolution of viral tolerance in a species-rich bat radiation.Cell Genom. 2024 Feb 14;4(2):100482. doi: 10.1016/j.xgen.2023.100482. Epub 2024 Jan 17. Cell Genom. 2024. PMID: 38237599 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
