All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues
- PMID: 18252004
- PMCID: PMC2267717
- DOI: 10.1186/1471-2164-9-67
All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues
Abstract
Background: The promoters of housekeeping genes are well-bound by RNA polymerase II (RNAP) in different tissues. Although the promoters of these genes are known to contain CpG islands, the specific DNA sequences that are associated with high RNAP binding to housekeeping promoters has not been described.
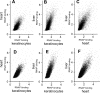
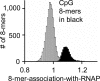
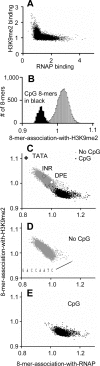
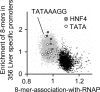
Results: ChIP-chip experiments from three mouse tissues, liver, heart ventricles, and primary keratinocytes, indicate that 94% of promoters have similar RNAP binding, ranging from well-bound to poorly-bound in all tissues. Using all 8-base pair long sequences as a test set, we have identified the DNA sequences that are enriched in promoters of housekeeping genes, focusing on those DNA sequences which are preferentially localized in the proximal promoter. We observe a bimodal distribution. Virtually all sequences enriched in promoters with high RNAP binding values contain a CpG dinucleotide. These results suggest that only transcription factor binding sites (TFBS) that contain the CpG dinucleotide are involved in RNAP binding to housekeeping promoters while TFBS that do not contain a CpG are involved in regulated promoter activity. Abundant 8-mers that are preferentially localized in the proximal promoters and exhibit the best enrichment in RNAP bound promoters are all variants of six known CpG-containing TFBS: ETS, NRF-1, BoxA, SP1, CRE, and E-Box. The frequency of these six DNA motifs can predict housekeeping promoters as accurately as the presence of a CpG island, suggesting that they are the structural elements critical for CpG island function. Experimental EMSA results demonstrate that methylation of the CpG in the ETS, NRF-1, and SP1 motifs prevent DNA binding in nuclear extracts in both keratinocytes and liver.
Conclusion: In general, TFBS that do not contain a CpG are involved in regulated gene expression while TFBS that contain a CpG are involved in constitutive gene expression with some CpG containing sequences also involved in inducible and tissue specific gene regulation. These TFBS are not bound when the CpG is methylated. Unmethylated CpG dinucleotides in the TFBS in CpG islands allow the transcription factors to find their binding sites which occur only in promoters, in turn localizing RNAP to promoters.
Figures


Similar articles
-
CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes.Proc Natl Acad Sci U S A. 2010 Nov 23;107(47):20311-6. doi: 10.1073/pnas.1008688107. Epub 2010 Nov 8. Proc Natl Acad Sci U S A. 2010. PMID: 21059933 Free PMC article.
-
Clusters of regulatory signals for RNA polymerase II transcription associated with Alu family repeats and CpG islands in human promoters.Genomics. 2004 May;83(5):873-82. doi: 10.1016/j.ygeno.2003.11.001. Genomics. 2004. PMID: 15081116
-
Overlapping ETS and CRE Motifs ((G/C)CGGAAGTGACGTCA) preferentially bound by GABPα and CREB proteins.G3 (Bethesda). 2012 Oct;2(10):1243-56. doi: 10.1534/g3.112.004002. Epub 2012 Oct 1. G3 (Bethesda). 2012. PMID: 23050235 Free PMC article.
-
Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation.Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194567. doi: 10.1016/j.bbagrm.2020.194567. Epub 2020 Apr 29. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32360393 Free PMC article. Review.
-
Sequence determinants, function, and evolution of CpG islands.Biochem Soc Trans. 2021 Jun 30;49(3):1109-1119. doi: 10.1042/BST20200695. Biochem Soc Trans. 2021. PMID: 34156435 Free PMC article. Review.
Cited by
-
High nucleosome occupancy is encoded at human regulatory sequences.PLoS One. 2010 Feb 9;5(2):e9129. doi: 10.1371/journal.pone.0009129. PLoS One. 2010. PMID: 20161746 Free PMC article.
-
Exploiting nucleotide composition to engineer promoters.PLoS One. 2011;6(5):e20136. doi: 10.1371/journal.pone.0020136. Epub 2011 May 18. PLoS One. 2011. PMID: 21625601 Free PMC article.
-
The bZIP mutant CEBPB (V285A) has sequence specific DNA binding propensities similar to CREB1.Biochim Biophys Acta Gene Regul Mech. 2019 Apr;1862(4):486-492. doi: 10.1016/j.bbagrm.2019.02.002. Epub 2019 Feb 27. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 30825655 Free PMC article.
-
Human and mouse switch-like genes share common transcriptional regulatory mechanisms for bimodality.BMC Genomics. 2008 Dec 23;9:628. doi: 10.1186/1471-2164-9-628. BMC Genomics. 2008. PMID: 19105848 Free PMC article.
-
Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells.RNA Biol. 2014;11(6):777-87. doi: 10.4161/rna.28828. Epub 2014 Apr 24. RNA Biol. 2014. PMID: 24824789 Free PMC article.
References
-
- Web site of the Charles Vinson laboratory http://home.ccr.cancer.gov/metabolism/vinson/vinsonccr.htm
-
- Maston GA, Evans SK, Green MR. Transcriptional Regulatory Elements in the Human Genome. Annu Rev Genomics Hum Genet. 2006. - PubMed
-
- Swartz MN, Trautner TA, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962;237:1961–1967. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

