The local and systemic inflammatory transcriptome after acute kidney injury
- PMID: 18235097
- PMCID: PMC2391061
- DOI: 10.1681/ASN.2007040469
The local and systemic inflammatory transcriptome after acute kidney injury
Abstract
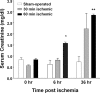
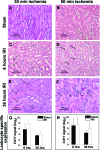
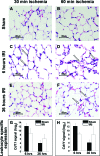
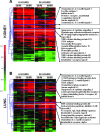
Studies in humans and animal models have demonstrated that acute kidney injury (AKI) has a significant effect on the function of extrarenal organs. The combination of AKI and lung dysfunction is associated with 80% mortality; the lung, because of its extensive capillary network, is a prime target for AKI-induced effects. The study presented here tested the hypothesis that AKI leads to a vigorous inflammatory response and produces distinct genomic signatures in the kidney and lung. In a murine model of ischemic AKI, prominent global transcriptomic changes and histologic injury in both kidney and lung tissues were identified. These changes were evident at both early (6 h) and late (36 h) timepoints after 60-min bilateral kidney ischemia and were more prominent than similar timepoints after sham surgery or 30 min of ischemia. The inflammatory transcriptome (109 genes) of both organs changed with marked similarity, including the innate immunity genes Cd14, Socs3, Saa3, Lcn2, and Il1r2. Functional genomic analysis of these genes suggested that IL-10 and IL-6 signaling was involved in the distant effects of local inflammation, and this was supported by increased serum levels of IL-10 and IL-6 after ischemia-reperfusion. In summary, this is the first comprehensive analysis of concomitant inflammation-associated transcriptional changes in the kidney and a remote organ during AKI. Functional genomic analysis identified potential mediators that connect local and systemic inflammation, suggesting that this type of analysis may be a useful discovery tool for novel biomarkers and therapeutic drug development.
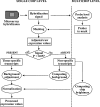
Figures

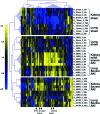
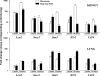
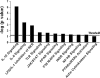
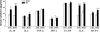
Similar articles
-
Splenectomy exacerbates lung injury after ischemic acute kidney injury in mice.Am J Physiol Renal Physiol. 2011 Oct;301(4):F907-16. doi: 10.1152/ajprenal.00107.2011. Epub 2011 Jun 15. Am J Physiol Renal Physiol. 2011. PMID: 21677145 Free PMC article.
-
Ischemic acute kidney injury induces a distant organ functional and genomic response distinguishable from bilateral nephrectomy.Am J Physiol Renal Physiol. 2007 Jul;293(1):F30-40. doi: 10.1152/ajprenal.00023.2007. Epub 2007 Feb 27. Am J Physiol Renal Physiol. 2007. PMID: 17327501
-
Concordant changes of plasma and kidney microRNA in the early stages of acute kidney injury: time course in a mouse model of bilateral renal ischemia-reperfusion.PLoS One. 2014 Apr 2;9(4):e93297. doi: 10.1371/journal.pone.0093297. eCollection 2014. PLoS One. 2014. PMID: 24695114 Free PMC article.
-
Role of Complement Properdin in Renal Ischemia-Reperfusion Injury.Curr Gene Ther. 2017;17(6):411-423. doi: 10.2174/1566523218666180214093043. Curr Gene Ther. 2017. PMID: 29446739 Review.
-
Mitochondria as mediators of systemic inflammation and organ cross talk in acute kidney injury.Am J Physiol Renal Physiol. 2022 Jun 1;322(6):F589-F596. doi: 10.1152/ajprenal.00372.2021. Epub 2022 Apr 4. Am J Physiol Renal Physiol. 2022. PMID: 35379000 Free PMC article. Review.
Cited by
-
Myocardial infarction causes inflammation and leukocyte recruitment at remote sites in the myocardium and in the renal glomerulus.Inflamm Res. 2013 May;62(5):515-25. doi: 10.1007/s00011-013-0605-4. Epub 2013 Mar 8. Inflamm Res. 2013. PMID: 23471223 Free PMC article.
-
Dendritic Cells as Sensors, Mediators, and Regulators of Ischemic Injury.Front Immunol. 2019 Oct 15;10:2418. doi: 10.3389/fimmu.2019.02418. eCollection 2019. Front Immunol. 2019. PMID: 31681306 Free PMC article. Review.
-
Renal Involvement in Multisystem Inflammatory Syndrome in Children: Not Only Acute Kidney Injury.Children (Basel). 2023 Oct 7;10(10):1661. doi: 10.3390/children10101661. Children (Basel). 2023. PMID: 37892324 Free PMC article.
-
The origin of plasma neutrophil gelatinase-associated lipocalin in cardiac surgery.BMC Nephrol. 2019 May 22;20(1):182. doi: 10.1186/s12882-019-1380-4. BMC Nephrol. 2019. PMID: 31113394 Free PMC article. Clinical Trial.
-
Meta-analysis of molecular response of kidney to ischemia reperfusion injury for the identification of new candidate genes.BMC Nephrol. 2013 Oct 24;14:231. doi: 10.1186/1471-2369-14-231. BMC Nephrol. 2013. PMID: 24152794 Free PMC article.
References
-
- Levy EM, Viscoli CM, Horwitz RI: The effect of acute renal failure on mortality. A cohort analysis. JAMA 275: 1489–1494, 1996 - PubMed
-
- Kramer AA, Postler G, Salhab KF, Mendez C, Carey LC, Rabb H: Renal ischemia/reperfusion leads to macrophage-mediated increase in pulmonary vascular permeability. Kidney Int 55: 2362–2367, 1999 - PubMed
-
- Kelly KJ: Distant effects of experimental renal ischemia/reperfusion injury. J Am Soc Nephrol 14: 1549–1558, 2003 - PubMed
-
- Rabb H, Wang Z, Postler G, Soleimani M: Possible molecular basis for changes in potassium handling in acute renal failure. Am J Kidney Dis 35: 871–877, 2000 - PubMed
-
- Zarbock A, Schmolke M, Spieker T, Jurk K, Van Aken H, Singbartl K: Acute uremia but not renal inflammation attenuates aseptic acute lung injury: a critical role for uremic neutrophils. J Am Soc Nephrol 17: 3124–3131, 2006 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

