MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis
- PMID: 18230805
- PMCID: PMC2259104
- DOI: 10.1101/gr.6587008
MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis
Abstract
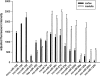
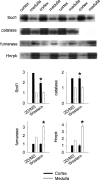
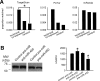
Mammalian genomes contain several hundred highly conserved genes encoding microRNAs. In silico analysis has predicted that a typical microRNA may regulate the expression of hundreds of target genes, suggesting miRNAs might have broad biological significance. A major challenge is to obtain experimental evidence for predicted microRNA-target pairs. We reasoned that reciprocal expression of a microRNA and a predicted target within a physiological context would support the presence and relevance of a microRNA-target pair. We used microRNA microarray and proteomic techniques to analyze the cortex and the medulla of rat kidneys. Of the 377 microRNAs analyzed, we identified 6 as enriched in the renal cortex and 11 in the renal medulla. From approximately 2100 detectable protein spots in two-dimensional gels, we identified 58 proteins as more abundant in the renal cortex and 72 in the renal medulla. The differential expression of several microRNAs and proteins was verified by real-time PCR and Western blot analyses, respectively. Several pairs of reciprocally expressed microRNAs and proteins were predicted to be microRNA-target pairs by TargetScan, PicTar, or miRanda. Seven pairs were predicted by two algorithms and two pairs by all three algorithms. The identification of reciprocal expression of microRNAs and their computationally predicted targets in the rat kidney provides a unique molecular basis for further exploring the biological role of microRNA. In addition, this study establishes a differential profile of microRNA expression between the renal cortex and the renal medulla and greatly expands the known differential proteome profiles between the two kidney regions.
Figures
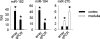
Similar articles
-
Identification of potential microRNA-target pairs associated with osteopetrosis by deep sequencing, iTRAQ proteomics and bioinformatics.Eur J Hum Genet. 2014 May;22(5):625-32. doi: 10.1038/ejhg.2013.221. Epub 2013 Oct 2. Eur J Hum Genet. 2014. PMID: 24084574 Free PMC article.
-
Differential expression of proteins in renal cortex and medulla: a proteomic approach.Kidney Int. 2002 Oct;62(4):1314-21. doi: 10.1111/j.1523-1755.2002.kid588.x. Kidney Int. 2002. PMID: 12234301
-
Mitochondrial proteomes of porcine kidney cortex and medulla: foundation for translational proteomics.Clin Exp Nephrol. 2016 Feb;20(1):39-49. doi: 10.1007/s10157-015-1135-x. Epub 2015 Jun 15. Clin Exp Nephrol. 2016. PMID: 26072732
-
A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions.RNA. 2008 Nov;14(11):2290-6. doi: 10.1261/rna.1188208. Epub 2008 Sep 23. RNA. 2008. PMID: 18812437 Free PMC article.
-
Systems biology and machine learning approaches identify drug targets in diabetic nephropathy.Sci Rep. 2021 Dec 6;11(1):23452. doi: 10.1038/s41598-021-02282-3. Sci Rep. 2021. PMID: 34873190 Free PMC article.
Cited by
-
let-7e downregulation characterizes early phase colonic adenoma in APCMin/+ mice and human FAP subjects.PLoS One. 2021 Apr 26;16(4):e0249238. doi: 10.1371/journal.pone.0249238. eCollection 2021. PLoS One. 2021. PMID: 33901189 Free PMC article.
-
Emerging Role of MiR-192-5p in Human Diseases.Front Pharmacol. 2021 Feb 23;12:614068. doi: 10.3389/fphar.2021.614068. eCollection 2021. Front Pharmacol. 2021. PMID: 33708127 Free PMC article. Review.
-
Gallic Acid Alleviates Glucolipotoxicity-Induced Nephropathy by miR-709-NFE2L2 Pathway in db/db Mice on a High-Fat Diet.J Agric Food Chem. 2024 Oct 4;72(41):22645-60. doi: 10.1021/acs.jafc.4c05898. Online ahead of print. J Agric Food Chem. 2024. PMID: 39365293 Free PMC article.
-
microRNA-802/Rnd3 pathway imposes on carcinogenesis and metastasis of fine particulate matter exposure.Oncotarget. 2016 Jun 7;7(23):35026-43. doi: 10.18632/oncotarget.9019. Oncotarget. 2016. PMID: 27144337 Free PMC article.
-
MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase β1.Nucleic Acids Res. 2013 Jan;41(2):1273-83. doi: 10.1093/nar/gks1228. Epub 2012 Dec 5. Nucleic Acids Res. 2013. PMID: 23221637 Free PMC article.
References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Arthur J.M., Thongboonkerd V., Scherzer J.A., Cai J., Pierce W.M., Klein J.B., Thongboonkerd V., Scherzer J.A., Cai J., Pierce W.M., Klein J.B., Scherzer J.A., Cai J., Pierce W.M., Klein J.B., Cai J., Pierce W.M., Klein J.B., Pierce W.M., Klein J.B., Klein J.B. Differential expression of proteins in renal cortex and medulla: a proteomic approach. Kidney Int. 2002;62:1314–1321. - PubMed
-
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Couzin J. Breakthrough of the year: Small RNAs make big splash. Science. 2002;298:2296–2297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
