Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice
- PMID: 18221561
- PMCID: PMC2267713
- DOI: 10.1186/1471-2164-9-44
Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice
Abstract
Background: Genes in the CCCH family encode zinc finger proteins containing the motif with three cysteines and one histidine residues. They have been known to play important roles in RNA processing as RNA-binding proteins in animals. To date, few plant CCCH proteins have been studied functionally.
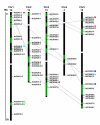
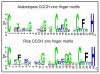
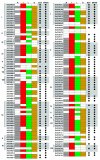
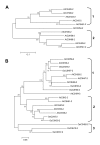
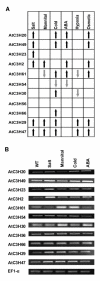
Results: In this study, a comprehensive computational analysis identified 68 and 67 CCCH family genes in Arabidopsis and rice, respectively. A complete overview of this gene family in Arabidopsis was presented, including the gene structures, phylogeny, protein motifs, and chromosome locations. In addition, a comparative analysis between these genes in Arabidopsis and rice was performed. These results revealed that the CCCH families in Arabidopsis and rice were divided into 11 and 8 subfamilies, respectively. The gene duplication contributed to the expansion of the CCCH gene family in Arabidopsis genome. Expression studies indicated that CCCH proteins exhibit a variety of expression patterns, suggesting diverse functions. Finally, evolutionary analysis showed that one subfamily is higher plant specific. The expression profile indicated that most members of this subfamily are regulated by abiotic or biotic stresses, suggesting that they could have an effective role in stress tolerance.
Conclusion: Our comparative genomics analysis of CCCH genes and encoded proteins in two model plant species provides the first step towards the functional dissection of this emerging family of potential RNA-binding proteins.
Figures





Similar articles
-
Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa).BMC Genomics. 2012 Jun 18;13:253. doi: 10.1186/1471-2164-13-253. BMC Genomics. 2012. PMID: 22708723 Free PMC article.
-
Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa.BMC Plant Biol. 2018 Dec 27;18(1):373. doi: 10.1186/s12870-018-1608-7. BMC Plant Biol. 2018. PMID: 30587139 Free PMC article.
-
Genome-wide analysis and identification of stress-responsive genes of the CCCH zinc finger family in Solanum lycopersicum.Mol Genet Genomics. 2014 Oct;289(5):965-79. doi: 10.1007/s00438-014-0861-1. Epub 2014 May 29. Mol Genet Genomics. 2014. PMID: 24870401
-
Hormone Regulation of CCCH Zinc Finger Proteins in Plants.Int J Mol Sci. 2022 Nov 18;23(22):14288. doi: 10.3390/ijms232214288. Int J Mol Sci. 2022. PMID: 36430765 Free PMC article. Review.
-
Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.).BMC Genomics. 2020 Oct 19;21(1):722. doi: 10.1186/s12864-020-07121-9. BMC Genomics. 2020. PMID: 33076828 Free PMC article. Review.
Cited by
-
Comparative Transcriptome and Expression Profiling of Resistant and Susceptible Banana Cultivars during Infection by Fusarium oxysporum.Int J Mol Sci. 2021 Mar 16;22(6):3002. doi: 10.3390/ijms22063002. Int J Mol Sci. 2021. PMID: 33809411 Free PMC article.
-
Comparative genome analysis of PHB gene family reveals deep evolutionary origins and diverse gene function.BMC Bioinformatics. 2010 Oct 7;11 Suppl 6(Suppl 6):S22. doi: 10.1186/1471-2105-11-S6-S22. BMC Bioinformatics. 2010. PMID: 20946606 Free PMC article.
-
RNA Binding Protein OsTZF7 Traffics Between the Nucleus and Processing Bodies/Stress Granules and Positively Regulates Drought Stress in Rice.Front Plant Sci. 2022 Feb 21;13:802337. doi: 10.3389/fpls.2022.802337. eCollection 2022. Front Plant Sci. 2022. PMID: 35265093 Free PMC article.
-
Zinc Finger Proteins in the Human Fungal Pathogen Cryptococcus neoformans.Int J Mol Sci. 2020 Feb 18;21(4):1361. doi: 10.3390/ijms21041361. Int J Mol Sci. 2020. PMID: 32085473 Free PMC article. Review.
-
PRR5, 7 and 9 positively modulate TOR signaling-mediated root cell proliferation by repressing TANDEM ZINC FINGER 1 in Arabidopsis.Nucleic Acids Res. 2019 Jun 4;47(10):5001-5015. doi: 10.1093/nar/gkz191. Nucleic Acids Res. 2019. PMID: 30892623 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

