Evolutionary dynamics of nematode operons: easy come, slow go
- PMID: 18218978
- PMCID: PMC2259105
- DOI: 10.1101/gr.7112608
Evolutionary dynamics of nematode operons: easy come, slow go
Abstract
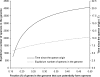
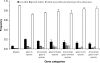
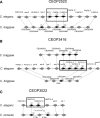
Operons are widespread in prokaryotes, but are uncommon in eukaryotes, except nematode worms, where approximately 15% of genes reside in over 1100 operons in the model organism Caenorhabditis elegans. It is unclear how operons have become abundant in nematode genomes. The "one-way street" hypothesis asserts that once formed by chance, operons are very difficult to break, because the breakage would leave downstream genes in an operon without a promoter, and hence, unexpressed. To test this hypothesis, we analyzed the presence and absence of C. elegans operons in Caenorhabditis briggsae, Caenorhabditis remanei, and Caenorhabditis brenneri, using Pristionchus pacificus and Brugia malayi as outgroups, and identified numerous operon gains and losses. Coupled with experimental examination of trans-splicing patterns, our comparative genomic analysis revealed diverse molecular mechanisms of operon losses, including inversion, insertion, and relocation, but the presence of internal promoters was not found to facilitate operon losses. In several cases, the data allowed inference of mechanisms by which downstream genes are expressed after operon breakage. We found that the rate of operon gain is approximately 3.3 times that of operon loss. Thus, the evolutionary dynamics of nematode operons is better described as "easy come, slow go," rather than a "one-way street." Based on a mathematic model of operon gains and losses and additional assumptions, we projected that the number of operons in C. elegans will continue to rise by 6%-18% in future evolution before reaching equilibrium between operon gains and losses.
Figures

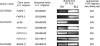

Similar articles
-
Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons.RNA. 2014 Sep;20(9):1386-97. doi: 10.1261/rna.041954.113. Epub 2014 Jul 11. RNA. 2014. PMID: 25015138 Free PMC article.
-
Operon structure and trans-splicing in the nematode Pristionchus pacificus.Mol Biol Evol. 2003 Dec;20(12):2097-103. doi: 10.1093/molbev/msg225. Epub 2003 Aug 29. Mol Biol Evol. 2003. PMID: 12949121
-
Operon conservation and the evolution of trans-splicing in the phylum Nematoda.PLoS Genet. 2006 Nov 24;2(11):e198. doi: 10.1371/journal.pgen.0020198. Epub 2006 Oct 9. PLoS Genet. 2006. PMID: 17121468 Free PMC article.
-
Trans-splicing and operons in C. elegans.WormBook. 2012 Nov 20:1-11. doi: 10.1895/wormbook.1.5.2. WormBook. 2012. PMID: 23175478 Free PMC article. Review.
-
Trans-splicing and operons.WormBook. 2005 Jun 25:1-9. doi: 10.1895/wormbook.1.5.1. WormBook. 2005. PMID: 18050426 Review.
Cited by
-
The evolutionary dynamics of operon distributions in eukaryote genomes.Genetics. 2010 Jun;185(2):685-93. doi: 10.1534/genetics.110.115766. Epub 2010 Apr 9. Genetics. 2010. PMID: 20382830 Free PMC article.
-
Large synteny blocks revealed between Caenorhabditis elegans and Caenorhabditis briggsae genomes using OrthoCluster.BMC Genomics. 2010 Sep 24;11:516. doi: 10.1186/1471-2164-11-516. BMC Genomics. 2010. PMID: 20868500 Free PMC article.
-
Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons.RNA. 2014 Sep;20(9):1386-97. doi: 10.1261/rna.041954.113. Epub 2014 Jul 11. RNA. 2014. PMID: 25015138 Free PMC article.
-
Genome annotation of Caenorhabditis briggsae by TEC-RED identifies new exons, paralogs, and conserved and novel operons.G3 (Bethesda). 2022 Jul 6;12(7):jkac101. doi: 10.1093/g3journal/jkac101. G3 (Bethesda). 2022. PMID: 35485953 Free PMC article.
-
Clusters of microRNAs emerge by new hairpins in existing transcripts.Nucleic Acids Res. 2013 Sep;41(16):7745-52. doi: 10.1093/nar/gkt534. Epub 2013 Jun 17. Nucleic Acids Res. 2013. PMID: 23775791 Free PMC article.
References
-
- Baird S.E., Sutherlin M.E., Emmons S.W., Sutherlin M.E., Emmons S.W., Emmons S.W. Reproductive isolation in Rhabditidae (Nematoda: Secernentea): Mechanisms that isolate six species of three genera. Evolution Int. J. Org. Evolution. 1992;46:585–594. - PubMed
-
- Blaxter M.L., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., De Ley P., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Garey J.R., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Liu L.X., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Scheldeman P., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vierstraete A., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Vanfleteren J.R., Mackey L.Y., Dorris M., Frisse L.M., Mackey L.Y., Dorris M., Frisse L.M., Dorris M., Frisse L.M., Frisse L.M., et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–75. - PubMed
-
- Blumenthal T. Gene clusters and polycistronic transcription in eukaryotes. Bioessays. 1998;20:480–487. - PubMed
-
- Blumenthal T. Operons in eukaryotes. Brief Funct. Genomic Proteomic. 2004;3:199–211. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
