Emerging roles of the SUMO pathway in mitosis
- PMID: 18218095
- PMCID: PMC2265688
- DOI: 10.1186/1747-1028-3-5
Emerging roles of the SUMO pathway in mitosis
Abstract
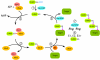
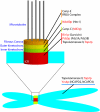
SUMO proteins are small ubiquitin-like modifiers found in all eukaryotes that become covalently conjugated to other cellular proteins. The SUMO conjugation pathway is biochemically similar to ubiquitin conjugation, although the enzymes within the pathway act exclusively on SUMO proteins. This post-translational modification controls many processes. Here, I will focus on evidence that SUMOylation plays a critical role(s) in mitosis: Early studies showed a genetic requirement for SUMO pathway components in the process of cell division, while later findings implicated SUMOylation in the control of mitotic chromosome structure, cell cycle progression, kinetochore function and cytokinesis. Recent insights into the targets of SUMOylation are likely to be extremely helpful in understanding each of these aspects. Finally, growing evidence suggests that SUMOylation is a downstream target of regulation through Ran, a small GTPase with important functions in both interphase nuclear trafficking and mitotic spindle assembly.
Figures
Similar articles
-
The SUMO Pathway in Mitosis.Adv Exp Med Biol. 2017;963:171-184. doi: 10.1007/978-3-319-50044-7_10. Adv Exp Med Biol. 2017. PMID: 28197912 Free PMC article. Review.
-
Cell cycle-dependent SUMO-1 conjugation to nuclear mitotic apparatus protein (NuMA).Biochem Biophys Res Commun. 2014 Jan 3;443(1):259-65. doi: 10.1016/j.bbrc.2013.11.107. Epub 2013 Dec 2. Biochem Biophys Res Commun. 2014. PMID: 24309115
-
RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on Borealin.Mol Biol Cell. 2009 Jan;20(1):410-8. doi: 10.1091/mbc.e08-05-0511. Epub 2008 Oct 22. Mol Biol Cell. 2009. PMID: 18946085 Free PMC article.
-
Converging Small Ubiquitin-like Modifier (SUMO) and Ubiquitin Signaling: Improved Methodology Identifies Co-modified Target Proteins.Mol Cell Proteomics. 2017 Dec;16(12):2281-2295. doi: 10.1074/mcp.TIR117.000152. Epub 2017 Sep 26. Mol Cell Proteomics. 2017. PMID: 28951443 Free PMC article.
-
New insights into the role of the small ubiquitin-like modifier (SUMO) in plants.Int Rev Cell Mol Biol. 2013;300:161-209. doi: 10.1016/B978-0-12-405210-9.00005-9. Int Rev Cell Mol Biol. 2013. PMID: 23273862 Review.
Cited by
-
Sumoylation and regulation of cardiac gene expression.Circ Res. 2010 Jul 9;107(1):19-29. doi: 10.1161/CIRCRESAHA.110.220491. Circ Res. 2010. PMID: 20616338 Free PMC article. Review.
-
Sumoylation and its regulation in testicular Sertoli cells.Biochem Biophys Res Commun. 2021 Nov 26;580:56-62. doi: 10.1016/j.bbrc.2021.09.066. Epub 2021 Sep 29. Biochem Biophys Res Commun. 2021. PMID: 34624570 Free PMC article.
-
SUMOlock reveals a more complete Aspergillus nidulans SUMOylome.Fungal Genet Biol. 2019 Jun;127:50-59. doi: 10.1016/j.fgb.2019.03.002. Epub 2019 Mar 5. Fungal Genet Biol. 2019. PMID: 30849444 Free PMC article.
-
Cross-talk between sumoylation and phosphorylation in mouse spermatocytes.Biochem Biophys Res Commun. 2017 Jun 3;487(3):640-645. doi: 10.1016/j.bbrc.2017.04.107. Epub 2017 Apr 20. Biochem Biophys Res Commun. 2017. PMID: 28435066 Free PMC article.
-
SUMO: a multifaceted modifier of chromatin structure and function.Dev Cell. 2013 Jan 14;24(1):1-12. doi: 10.1016/j.devcel.2012.11.020. Dev Cell. 2013. PMID: 23328396 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

