High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes
- PMID: 18212089
- PMCID: PMC2279250
- DOI: 10.1101/gr.072942.107
High-throughput biochemical analysis of in vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes
Erratum in
- Genome Res. 2009 Apr;19(4):690. Fairbrother, William [corrected to Fairbrother, William G]
Abstract
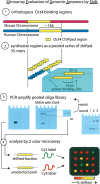
The transcription factor POU5F1 is a key regulator of embryonic stem (ES) cell pluripotency and a known oncoprotein. We have developed a novel high-throughput binding assay called MEGAshift (microarray evaluation of genomic aptamers by shift) that we use to pinpoint the exact location, affinity, and stoichiometry of the DNA-protein complexes identified by chromatin immunoprecipitation studies. We consider all genomic regions identified as POU5F1-ChIP-enriched in both human and mouse. Compared with regions that are ChIP-enriched in a single species, we find these regions more likely to be near actively transcribed genes in ES cells. We resynthesize these genomic regions as a pool of tiled 35-mers. This oligonucleotide pool is then assayed for binding to recombinant POU5F1 by gel shift. The degree of binding for each oligonucleotide is accurately measured on a custom oligonucleotide microarray. We explore the relationship between experimentally determined and computationally predicted binding strengths, find many novel functional combinations of POU5F1 half sites, and demonstrate efficient motif discovery by incorporating binding information into a motif finding algorithm. In addition to further refining location studies for transcription factors, this method holds promise for the high-throughput screening of promoters, SNP regions, and epigenetic modifications for factor binding.
Figures

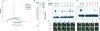

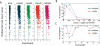

Similar articles
-
Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data.BMC Genomics. 2008 Jun 3;9:269. doi: 10.1186/1471-2164-9-269. BMC Genomics. 2008. PMID: 18522731 Free PMC article.
-
Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET.Genome Biol. 2008;9(8):R126. doi: 10.1186/gb-2008-9-8-r126. Epub 2008 Aug 13. Genome Biol. 2008. PMID: 18700969 Free PMC article.
-
Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1.Nat Cell Biol. 2006 Oct;8(10):1114-23. doi: 10.1038/ncb1481. Epub 2006 Sep 17. Nat Cell Biol. 2006. PMID: 16980957
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
-
OCT4: dynamic DNA binding pioneers stem cell pluripotency.Biochim Biophys Acta. 2014 Mar;1839(3):138-54. doi: 10.1016/j.bbagrm.2013.10.001. Epub 2013 Oct 18. Biochim Biophys Acta. 2014. PMID: 24145198 Review.
Cited by
-
RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections.Nucleic Acids Res. 2017 Jul 27;45(13):e119. doi: 10.1093/nar/gkx314. Nucleic Acids Res. 2017. PMID: 28591841 Free PMC article.
-
Targeting TBP-Associated Factors in Ovarian Cancer.Front Oncol. 2014 Mar 11;4:45. doi: 10.3389/fonc.2014.00045. eCollection 2014. Front Oncol. 2014. PMID: 24653979 Free PMC article. Review.
-
Dry and wet approaches for genome-wide functional annotation of conventional and unconventional transcriptional activators.Comput Struct Biotechnol J. 2016 Jun 29;14:262-70. doi: 10.1016/j.csbj.2016.06.004. eCollection 2016. Comput Struct Biotechnol J. 2016. PMID: 27453771 Free PMC article. Review.
-
High-throughput binding analysis determines the binding specificity of ASF/SF2 on alternatively spliced human pre-mRNAs.Comb Chem High Throughput Screen. 2010 Mar;13(3):242-52. doi: 10.2174/138620710790980522. Comb Chem High Throughput Screen. 2010. PMID: 20015017 Free PMC article.
-
Using protein-binding microarrays to study transcription factor specificity: homologs, isoforms and complexes.Brief Funct Genomics. 2015 Jan;14(1):17-29. doi: 10.1093/bfgp/elu046. Epub 2014 Nov 26. Brief Funct Genomics. 2015. PMID: 25431149 Free PMC article. Review.
References
-
- Ambrosetti D.C., Basilico C., Dailey L., Basilico C., Dailey L., Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 1997;17:6321–6329. - PMC - PubMed
-
- Atlasi Y., Mowla S.J., Ziaee S.A., Bahrami A.R., Mowla S.J., Ziaee S.A., Bahrami A.R., Ziaee S.A., Bahrami A.R., Bahrami A.R. OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int. J. Cancer. 2007;120:1598–1602. - PubMed
-
- Botquin V., Hess H., Fuhrmann G., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R., Hess H., Fuhrmann G., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R., Fuhrmann G., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R., Gross M.K., Vriend G., Scholer H.R., Vriend G., Scholer H.R., Scholer H.R. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes & Dev. 1998;12:2073–2090. - PMC - PubMed
-
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. - PMC - PubMed
-
- Buck M.J., Lieb J.D., Lieb J.D. ChIP-chip: Considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
