A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes
- PMID: 1820208
- PMCID: PMC5952205
A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes
Abstract
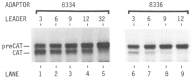
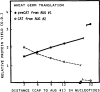
The functional consequences of unusually short 5' noncoding sequences on eukaryotic mRNAs are explored here by using an in vitro transcription and translation system. As the distance of the first AUG codon from the m7G cap was decreased from 32 to 3 nucleotides, the yield of protein initiated from the first AUG codon progressively decreased, with a corresponding increase in initiation from the second AUG codon. The leakiness attributable to a too-short leader sequence was offset, however, by introducing secondary structure downstream from the first AUG codon.
Figures

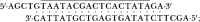
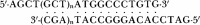
Similar articles
-
Effects of long 5' leader sequences on initiation by eukaryotic ribosomes in vitro.Gene Expr. 1991 May;1(2):117-25. Gene Expr. 1991. PMID: 1820209 Free PMC article.
-
Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes.Proc Natl Acad Sci U S A. 1990 Nov;87(21):8301-5. doi: 10.1073/pnas.87.21.8301. Proc Natl Acad Sci U S A. 1990. PMID: 2236042 Free PMC article.
-
The human immunodeficiency virus type 1 5' packaging signal structure affects translation but does not function as an internal ribosome entry site structure.J Virol. 1996 Feb;70(2):944-51. doi: 10.1128/JVI.70.2.944-951.1996. J Virol. 1996. PMID: 8551634 Free PMC article.
-
How do eucaryotic ribosomes select initiation regions in messenger RNA?Cell. 1978 Dec;15(4):1109-23. doi: 10.1016/0092-8674(78)90039-9. Cell. 1978. PMID: 215319 Review. No abstract available.
-
Cap-dependent, scanning-free translation initiation mechanisms.Biochim Biophys Acta. 2015 Nov;1849(11):1313-8. doi: 10.1016/j.bbagrm.2015.09.006. Epub 2015 Sep 14. Biochim Biophys Acta. 2015. PMID: 26381322 Review.
Cited by
-
Non-coding RNA: what is functional and what is junk?Front Genet. 2015 Jan 26;6:2. doi: 10.3389/fgene.2015.00002. eCollection 2015. Front Genet. 2015. PMID: 25674102 Free PMC article.
-
Non-canonical translation in RNA viruses.J Gen Virol. 2012 Jul;93(Pt 7):1385-1409. doi: 10.1099/vir.0.042499-0. Epub 2012 Apr 25. J Gen Virol. 2012. PMID: 22535777 Free PMC article. Review.
-
Preinitiation Complex Loading onto mRNAs with Long versus Short 5' TLs.Int J Mol Sci. 2022 Nov 2;23(21):13369. doi: 10.3390/ijms232113369. Int J Mol Sci. 2022. PMID: 36362157 Free PMC article.
-
mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5.Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9789-93. doi: 10.1073/pnas.88.21.9789. Proc Natl Acad Sci U S A. 1991. PMID: 1946403 Free PMC article.
-
Upstream AUGs in embryonic proinsulin mRNA control its low translation level.EMBO J. 2003 Oct 15;22(20):5582-92. doi: 10.1093/emboj/cdg515. EMBO J. 2003. PMID: 14532130 Free PMC article.
References
-
- Cate R. L., Mattaliano R. J., Hession C., Tizard R., Farber N. M., Cheung A., Ninfa E. G., Frey A. Z., Gash D. J., Chow E. P., Fisher R. A., Bertonis J. M., Torres G., Wallner B. P., Ramachandran K. L., Ragin R. C., Manganaro T. F., MacLaughlin D. T., and Donahoe P. K. (1986), Cell 45, 685–698. - PubMed
-
- Dasgupta R., Shih D. S., Saris C., and Kaesberg P. (1975), Nature 256, 624–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
